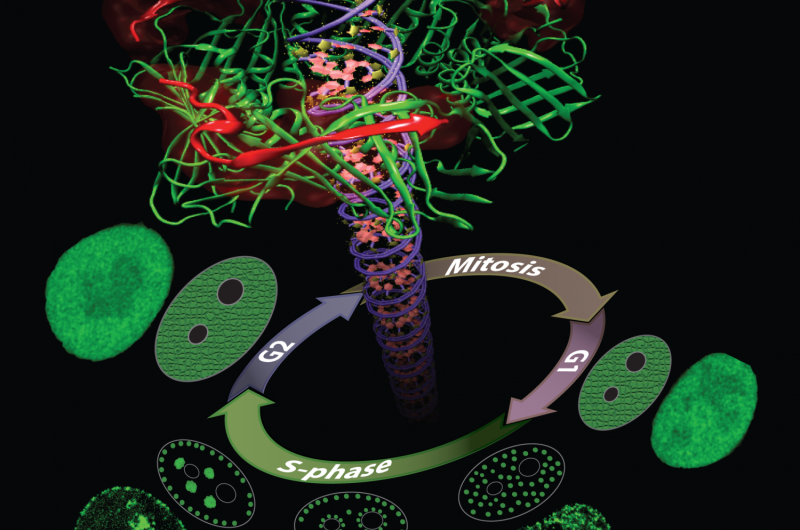
Proliferating Cell Nuclear Antigen (PCNA) is a key protein in DNA replication and repair. During the DNA synthesis phase (S-phase) PCNA is recruited to sub-nuclear sites of DNA replication forming distinctive patterns over time that characterize different S-phase stages. Simulations on TACC's Stampede supercomputer helped researchers understand these processes. Credit: Henry Herce
An important factor in fighting cancer is the speed at which the disease can be identified, diagnosed and treated.
The current standard involves a patient feeling ill or a physician seeing signs of a tumor. These indicators lead to more precise diagnoses via blood tests, x-rays or MRI imaging. But once the disease is far enough along to be noticeable, the cancer has often spread.
In the future, though, it may be possible to diagnose cancer much earlier using more sensitive body scans, new types of biomarker tests, and even nano-sensors working in the bloodstream.
Experimenting with these techniques in cancer patients or healthy individuals is difficult and potentially unethical. But scientists can test these technologies virtually using supercomputers to simulate the dynamics of cells and tissues.
Building a better breast cancer early detection system
Manual breast exams and mammograms are currently the most effective and widely used techniques for early detection of breast cancer. Unfortunately, manual breast exams are limited in their ability to detect tumors since they only produce local information about the site where the force is applied.
Mammograms (breast x-rays), on the other hand, are more accurate, but expose patients to radiation. Importantly, they do not quantify tissue stiffness, an identifying characteristic of breast tumors. They also produce many false positives, resulting in painful biopsies.
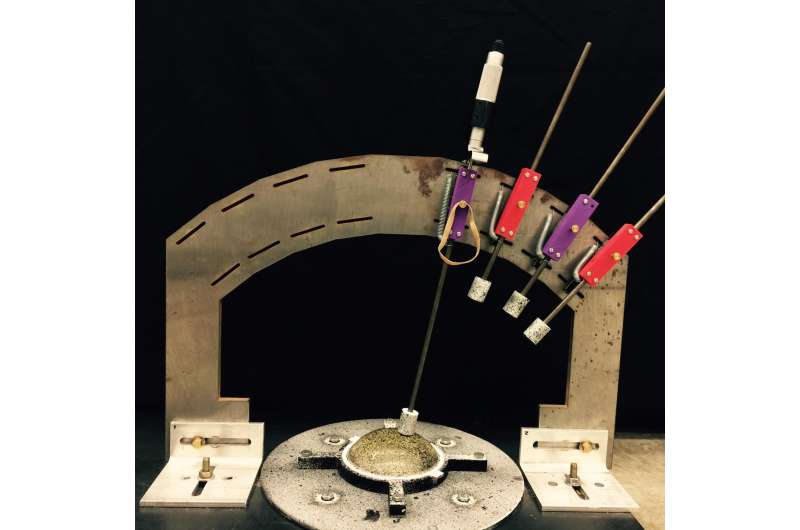
Lorraine Olson, a professor of mechanical engineering at Rose-Hulman Institute of Technology, is collaborating with colleagues Robert Throne of Electrical and Computer Engineering and Adam Nolte of Chemical Engineering to develop an electro-mechanical device that gently indents breast tissue in various locations and records the tissue surface deflections. This data is then converted into detailed 3-D maps of breast tissue stiffness, which can then be used to identify suspicious (stiffer) sites for further testing.
"The research takes an approach to early detection of breast cancer that utilizes a fundamental mechanical difference between cancerous and noncancerous tissue," Olson said. "Although this stiffness difference is the basis of manual breast exams, it has not been systematically investigated from an engineering point of view."
Olson and her team's approach to determining the relationship between stiffness and interior mapping involves a combination of finite element methods—a numerical method for solving problems in engineering and mathematical physics—and genetic algorithms—a method for solving optimization problems based on natural selection.
Paired together, they can map the distribution of stiffness in a given tissue and systematically use "guesses and checks" to find which tissue stiffness map best models the response they actually see in testing.
The process involves thousands of these "guesses" and therefore requires powerful supercomputers like Stampede at the Texas Advanced Computing Center (TACC), one of the most powerful in the world.
After numerous computer studies, the team has begun to experimentally validate this model using gelatin tissue phantoms (similar to Jell-O) with and without stiffer "tumors." They have been running indentation experiments to measure surface displacements on the tissue and identify tumor locations. They presented their work, which is supported by the National Science Foundation, at the 2016 Inverse Problems Symposium.
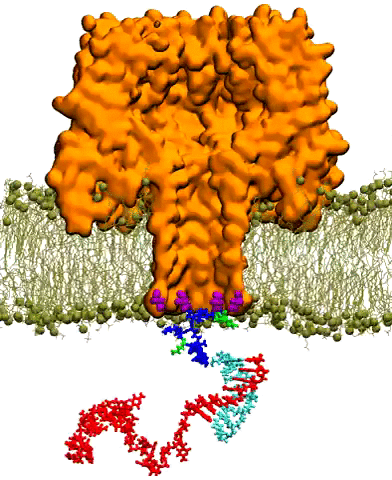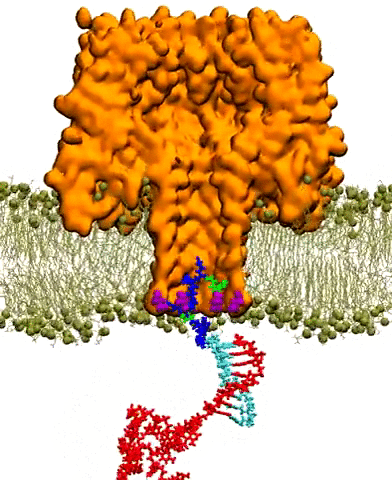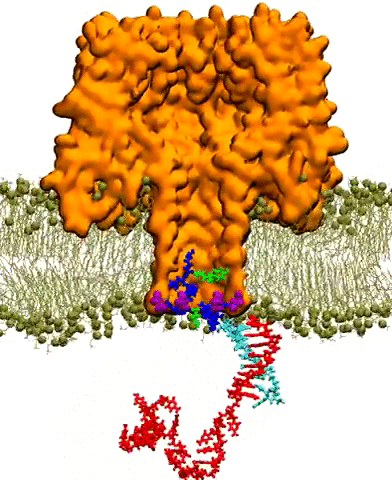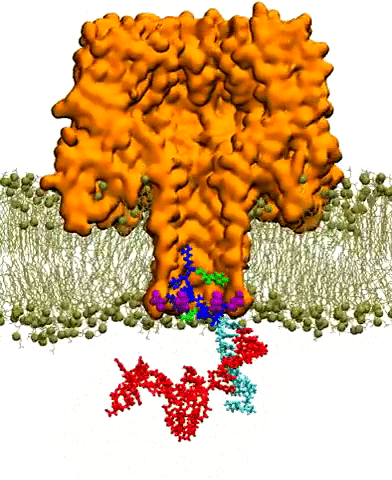
Molecular dynamics simulations on Stampede reproduced the capture of nanocarrier-DNA complex by a mutant alpha-hemolysin pore embedded in lipid bilayer. This video depicts ~35 nanoseconds of simulation time. The protein is shown in orange cutaway, the membrane in ochre lines and spheres, and the nanocarrier-DNA complex in licorice: DNA in red, proteo-nucleic acids (PNA) in teal, and polycationic peptide tag in blue and green. Credit: Kai Tian, Karl Decker, Aleksei Aksimentiev, and Li-Qun Gu, University of Missouri, University of Illinois at Urbana-Champaign
"This system has the potential to significantly increase the early detection of breast cancer with no unnecessary radiation, essentially no risk, and with little additional cost," Olson said.
Designing nanoscale dna-readers
Olson, Throne and Nolte's electromechanical technique works on the surface of the body, but an emerging class of nano-scale sensors aims to diagnose cancer from within the body.
Nanosensors must be small and sensitive, targeting specific biomarkers that may indicate the presence of cancer. They must also be able to communicate that information to an outside observer. Scientists and sci-fi authors have long predicted the rise of nanosensors, but only recently has it become feasible to engineer such technologies.
A number of scientists have been using TACC's supercomputers to investigate aspects of this problem. One such researcher is Aleksei Aksimentiev, a professor of biological physics at the University of Illinois, Urbana-Champaign. Aksimentiev focuses on creating silicon nanopore devices that can sequence DNA inside the body to detect the telltale signs of cancer or other diseases.
A nanopore is essentially a tiny hole in a very thin membrane, through which an even smaller particle, like DNA, can pass. In addition to being precisely shaped, it must be able to attract the right molecules and induce them to pass through the pore so they can be genetically sequenced and identified.
Writing in ACS Nano in December 2016, Aksimentiev and bioengineering professor Li-Qun (Andrew) Gu from the University of Missouri's Dalton Cardiovascular Research Center described efforts to detect genetic biomarkers using nanopores and synthetic nanocarriers. The nanocarriers selectively bind to target biomolecules, and increase their response to the electric field gradient generated by the nanopore, essentially forcing them through the hole.
The researchers showed that modestly charged nanocarriers can be used to detect and capture DNA or RNA molecules of any length or secondary structure. Such selective, molecular detection technologies would greatly improve the real-time analysis of complex clinical samples for cancer detection and other diseases.
Aksimentiev used TACC's Stampede supercomputer, as well as Blue Waters at the National Center for Supercomputing Applications, to design and virtually test the behavior of these nanopores systems.
"In the development of nanosensors, such as the nanopore single-molecule sensor for genetic diagnosis of cancer, we can experimentally discover various clinically useful phenomena at the nanometer scale. But our collaborator, Dr. Aksimentiev can utilize their superior computational power to accurately dig out the molecular mechanisms behind these experimental observations," said Gu. "These new nano-mechanisms can guide the design of a new generation of nanopore sensors for genetic marker-based cancer diagnostics, which we believe will play an important role in precision oncology."
(This work was supported by grants from the National Institutes of Health (R01-GM079613, R01-GM114204). The simulation studies led to the development of a patent, which was published in May 2017.)
Diagnosing biomarkers in the bloodstream with a microscopic lab-on-a-chip
Researchers from Lamar University, including Tao Wei, Ian Lian and Yu-Hwa Lo, are exploring a different approach to nanoscale cancer diagnostics. In place of nanopores, they are using lab-on-a-chip techniques invented by Lo's group that can recognize short nucleic acid fragments—which act as biomarkers for diseases—in the bloodstream.
System used to collect data from tissue phantoms for a new breast cancer diagnostic system. Data collected by the device is computationally modeled to identify possible tumors. Credit: Lorraine Olson, Robert Throne, Adam Nolte, Rose-Hulman Institute of Technology
The detection is based on the hybridization of nucleic acid fragments grafted on the substrate surface inside an evaporating droplet. Hybridization occurs when nucleic acids form hydrogen bonds. This process is influenced by the molecular geometry of the nucleic acid fragments that spontaneously self-organize into an ordered structure on the surface, also known as self-assembling monolayers (SAMs).
The sensitivity of such biochips is dependent on the degree to which the target material can bind to the surface of the biochip. Various factors affect the binding processes, including the structure of SAMs surface, the ion strength, the target DNA concentration and the surface packing density.
Wei and his collaborators used Stampede to perform molecular dynamics simulations and free energy computations to study the DNA hybridization process in detail.
"Our unique contribution is the use of atomistic simulation to design the surface and optimize the conditions to enhance hybridization efficiency in order to increase the detection resolution," said Wei. "The research will facilitate the development of new biodevices based on nucleic acid hybridization."
The team presented their work at the 2016 Annual Meeting of the American Institute of Chemical Engineers and are working to design and test experimental sensors capable of detecting nucleic acid biomarkers.
Sensors that enter nuclei and report on dna repair
Henry Herce, a research scientist at Rensselaer Polytechnic Institute (currently at Dana-Farber Cancer Institute) takes yet another tact to solve the problem of cancer detection in living cells. His research focuses on developing molecules that can enter the nucleus of a cell and bind to a specific protein there known as the Proliferating Cell Nuclear Antigen (PCNA).
PCNA, nicknamed the "ring master of the genome," is one of the key proteins in DNA replication and repair. DNA replication is essential for the survival, growth and spread of cancer, making it an intriguing target for tumor labeling and inhibition.
In a series of publications in the Journal of the American Chemical Society (2014) and Nucleus (2014) enabled by Stampede, Herce and his collaborators described the creation of a novel peptide - a chain of several amino acids—that can enter the cells of an organism, separate from its carrier molecule and bind to PCNA. They also uncovered the nature of the interactions between the peptides and their PCNA targets in atomistic detail.
Finally, they showed that both replication and repair sites can be directly labeled in live cells—the first cell-permeable peptide marker for these two fundamental processes—and introduced a PCNA staining method that causes the target molecule to light up so they can assess its distribution.
The simulations complemented experimental results and offered insights into how chemical modifications to the peptide's design can enhance its efficiency, increase its stability, and allow for intracellular delivery.
"The discovery represents a versatile tool for instantaneously labeling repair and replication processes in fixed and live cells," Herce said. "Down the road, it could lead to a new tool for early cancer detection."
More information: Kai Tian et al, Interference-Free Detection of Genetic Biomarkers Using Synthetic Dipole-Facilitated Nanopore Dielectrophoresis, ACS Nano (2016). DOI: 10.1021/acsnano.6b07570
Journal information: ACS Nano , Journal of the American Chemical Society
Provided by University of Texas at Austin