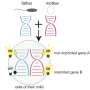
Gene variant activity is surprisingly variable between tissues

Every gene in almost every cell of the body is present in two variants called alleles—one from the mother, the other one from the father. In most cases, both alleles are active and transcribed by the cells into RNA. However, for a few genes, only one allele is expressed, while the other one is silenced. The decision to shut down either the maternal or the paternal version occurs early in embryonic development—it was long believed that the pattern of active alleles is nearly homogeneous in the various tissues of the organism.
The new study uncovers a different picture. By performing the first comprehensive analysis of all active alleles in 23 tissues and developmental stages of mice, the team of scientists discovered that each tissue shows a specific distribution of active alleles.
For their experiments, the researchers created hybrids of two genetically distinct mouse strains with a fully sequenced genome, allowing gene variants to be clearly assigned to the maternal or paternal allele. To facilitate the analysis, the team developed a user-friendly program called Allelome.PRO, that can easily be applied to similar datasets in mice and other species, a valuable tool for the community to investigate regulation of allele activity. By using this tool to analyze their data, the scientists catalogued active alleles in a comprehensive set of mouse tissues, or the mouse "Allelome," to gain insight into how this differential gene activity is regulated.
The scientists found that both genetic and epigenetic differences between the maternal and paternal allele contributed to the observed tissue-specific activity patterns. "Our results indicate that a large part of those patterns are induced by so-called 'enhancers,'" co-author Quanah Hudson explains. "Enhancers are DNA regions that are often located at quite some distance from the observed allele, but nevertheless have a direct influence on their activity."
Co-author Florian Pauler says, "This study reveals for the first time a comprehensive picture of all active alleles in different tissues—we have uncovered the first complete allelome. This is not only valuable for understanding basic biological functions, but will also help investigating diseases that involve defective gene regulators."
Some of the genes that contributed to the tissue-specific activity patterns were located on the X chromosome and escaped so-called X-chromosome inactivation, in which one of the two X chromosomes in females gets shut down. Previously, it was reported that around 3 percent of X-chromosomal genes in mice and 15 percent in humans escape inactivation. However, this study revealed that mice are more similar to humans than previously thought, with an average of around 10 percent of active genes escaping X-inactivation per tissue. By examining a broad range of organs, the researchers showed that the number of escapers varies dramatically between tissues. Most strikingly, muscle showed a surprisingly high rate of escapers, with over 50 percent of active genes escaping X chromosome activation, a result that may be relevant to some diseases of the muscle.
Finally, the allelome offers a near-complete picture of "genomic imprinting," the process that leads to epigenetic silencing of either the maternal or paternal allele that is initiated by an epigenetic mark placed in either the egg or sperm. Previously, it was reported that approximately 100 genes were subject to imprinted silencing—but in many cases, the tissue specificity was not known. This study led to the discovery of 18 new imprinted genes, validated some known genes and resolved the disputed status of some others to provide a gold standard list of 93 imprinted genes in mouse.
The scientists found that those new genes were located near other imprinted genes, indicating that they were co-regulated. Interestingly, this study demonstrated that Igfr2, the first imprinted gene discovered by Denise Barlow in 1991, is surrounding by a large cluster of imprinted genes that extend over 10 percent of the chromosome, making it the largest co-regulated domain in the genome outside of the X chromosome. Fittingly, her lab found the first imprinted gene and discovered the first imprinted non-coding RNA controlling imprinted silencing.
More information: Daniel Andergassen et al, Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression, eLife (2017). DOI: 10.7554/eLife.25125