A small drop of blood for an ocean of information
Patient response to treatment—especially personalised medicine—can be very difficult to predict. To overcome this issue, the CHEMOS project has been advancing a new method for screening thousands of single-cell drug responses from small blood samples.
The new method, called pharmacoscopy, combines automated microscopy and single-cell image analysis to provide over 20 million cells worth of data. Thanks to the I-FIVE project, which ran from 2010 to 2015, it had successfully been used to screen for novel anti-viral or immune modulating drugs. The project team had also demonstrated that the approach could help haema-oncologists to make therapeutic decisions in a concrete clinical setting using primary myelofibrosis and lymphoma as test diseases.
With CHEMOS (Chemical Haematology: breaking resistance of haematological malignancies through personalised drug trials), Prof. Dr. Giulio Superti-Furga and his team aimed to bring their screening method closer to market: the project looks to obtain clinical data through retrospective trials and use results to attract potential investors.
Prof. Superti-Furga agreed to discuss the project results ahead of its completion in September.
How would you explain the fact that personalised medicine for blood cancer has so far failed to deliver on its promises?
For the most part, personalised medicine for both blood and solid cancers relies on functional screening technology that focuses on the average characteristics of response to drugs. This generalisation does not discriminate against target effects—whereas we believe that such discrimination is very important in predicting patient response.
Besides, prior functional assays have measured early- or late-stage cytotoxicity using readouts such as global ATP levels, which have not provided robust enough responses to be used routinely within a clinical setting. On top of that, these assays require a lot of material to get above detection limit thresholds, and assays such as automated flow cytometry pose the additional problem that they require a hands-on component. Of course, these functional assays have paved the way for our research. But work on these approaches focused on patient stratification, ex vivo response profiling, drug discovery and mechanism of action elucidation: they have yet to become clinical routine.
Another issue lies in the fact that genetics—which really has shown the path towards personalised treatment of solid tumours—may prove more difficult to apply to haematological malignancies due to the diversity of clonal evolution during cancer progression and treatment. We find that our work combines very well with genetics, be it focused genetics and or more global genetic approaches, and should lead to mechanistic insights and new targetable pathways.
How does your new screening method provide a solution to these problems?
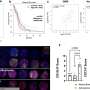
We believe that discrimination of drugs' effect on healthy versus cancer cells—an idea that is lost upon averaging a read-out in prior assays—is key to predicting response. Therefore, in our programme, we use high-content microscopy to determine—at single-cell resolution—the effects of drugs on each individual cell. In most cases, these effects imply cell death, as determined by nuclear disintegration of each cell measured in microscopy images.
The cancer cell phenotypes can be separated from the healthy cell phenotypes using fluorescent antibodies against diagnostic markers, just like a pathologist would do it. By performing this assay at single-cell resolution and on a large-scale, automated manner, each cell becomes an assay. This enables us to gather a differential cell response and to track precisely the drugs that kill cancer cells while leaving all other healthy cell material viable.
We can do this over thousands of cells per drug, and hundreds of thousands of drugs per patient sample. This all results in very robust measurements with dramatic statistical power, gathered with little human intervention, as the setup can be fully automated and the need for material is minimal. The images are also unique in that they provide a treasure-trove of data for us to dig into.
How do you proceed exactly?
Each well, part of a 384-well screening plate, is coated with a drug. Patient cells are put into each plate, and we create a monolayer of the cells which we view under an automated confocal microscope. This results in about 2 000 images per 384-well plate, and a total of 20 million cells worth of data. These images are then placed into an image analysis pipeline that extracts features of interest.
What would you say makes the project outcomes so innovative?
We found a solution where a 384-well plate doesn't imply 384 tests, but a single one that contains the data from approximately 20 million cells. This is really the basis, we think, of a 'big data' era for medicine, and we may just be scratching the surface of what data is contained in these images, and what part of that data can be translated. This is a major finding. From a more conceptual point of view, we found that some 10 % of commonly used therapeutics bear the property for modulating the immune system.
Which diseases could be targeted by this method? How so?
We have focused on haematological malignancies because of the ease at which samples can be extracted from patients during routine visits (much of the sample we get is left over from routine pathology). We have also started to look at other types of diseases, such as autoinflammatory diseases, starting with rheumatoid arthritis, albeit for other types of personalised medicine programmes.
What has been the feedback from potential investors so far?
Feedback has been very positive from both business and strategic investors, as well as government-backed programmes here in Austria.
How do you plan to get CHEMOS results to the market?
We have founded a company, Allcyte, here in Vienna that will focus on bringing this technology to market.
More information: Project page: cordis.europa.eu/project/rcn/205773