Antibiotic-resistant plasmids flourish in hospital plumbing
Antibiotic-resistant organisms can be found in multiple locations in a hospital - on countertops and doorknobs, on computers and in sinks, and even inside the plumbing. To better understand how these organisms spread, investigators at the National Institutes of Health (NIH) in Bethesda, Maryland, recently collected samples from pipes beneath the hospital's intensive care unit and from outside manholes draining hospital wastewater. They conducted whole-genome analyses on the samples to study the bacterial plasmids, or rings of DNA, that can confer resistance to antibiotics.
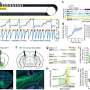
The majority of samples they studied from the pipes and sewers tested positive for bacterial plasmids that confer resistance to carbapenems, the researchers report this week in mBio. Carbapenems are "last-resort" antibiotics given to hospital patients who develop infections from pathogens that are multidrug-resistant. The new findings add to a growing body of evidence suggesting that the conduits of hospital wastewater serve as a vast and resilient reservoir for plasmids that can confer the genes responsible for antibiotic resistance.
Some scientists suggest that these populations flourish in waste because of the common use of strong antibiotics in hospitals, which leads to an uptick in antibiotic-resistant microbes in the sewers. Microorganisms compete for survival in the environment, says NIH microbiologist Karen Frank, who co-led the current study. "The bacteria fight with each other and plasmids can carry genes that help them survive," she says. As part of a complex bacterial community, they can transfer the plasmids carrying resistance genes to each other. That lateral gene transfer means bacteria can gain resistance, even without exposure to the antibiotics.
Frank and her collaborators compared their data to five years' worth of patient data and samples collected from sinks and other high-touch areas, like countertops, door knobs, and computers. Remarkably, the high prevalence of carbapenem-resistant plasmids in the pipes and sewers wasn't observed in parts of the hospital to which patients had access. Of 217 samples analyzed from high-touch surfaces, only three (1.4 percent) tested positive for carbapenem-resistant organisms. Similarly, of 340 samples collected from drains, only 11 (3.2 percent) were positive.
That comparison suggests that surveillance efforts to watch for resistant organisms are successful in minimizing patient infections, even so close to a reservoir of resistant bacteria, says Frank. "If you're tracking resistant bacteria, you might be able to prevent more infections in patients." The comparison also raises an important question, she adds. "How much should we care that there are a bunch of plasmids down in the wastewater system if they're not infecting our patients?"
Understanding the plasmid exchange and when the plasmids get into the pathogens that infect our patients, she says, could help hospitals improve their monitoring of resistance-conferring genes: "In the big picture, the concern is the spread of these resistant organisms worldwide and some regions of the world are not tracking the spread of the hospital isolates."
In 2011, the NIHCC experienced a cluster of infections of carbapenem-resistant Klebsiella pneumoniae. Using whole genome sequencing, researchers traced the chains of transmission. That analysis was led by Frank's co-study leaders, epidemiologist Tara Palmore, also at the NIH Clinical Center, and geneticist Julie Segre, at the National Human Genome Research Institute. The analysis published in mBio, says Segre, uses newer DNA sequencing technology that lets researchers compare microbes from patients and the environment.
Palmore says that the 2011 NIHCC outbreak led the hospital to institute additional surveillance, including increased monitoring of high-risk patients and regular sampling of the hospital environment. By knowing where the resistance-conferring genes hide, she says, researchers have a better chance of keeping them away from patients.