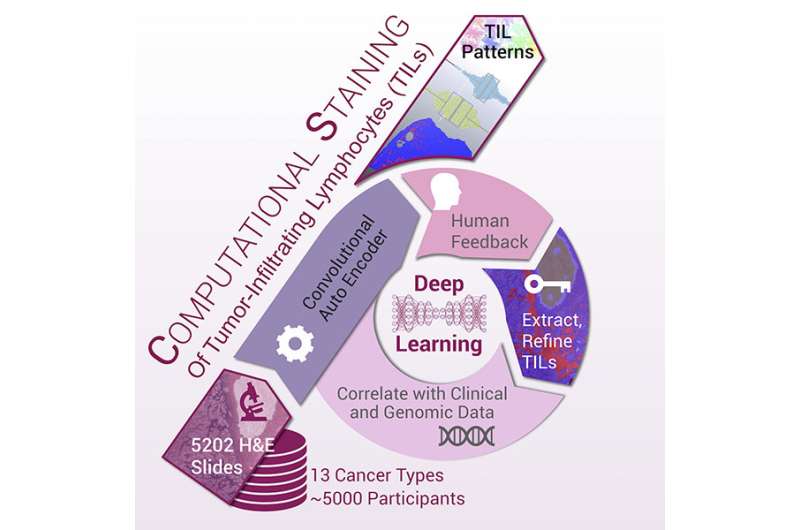
An illustration depicting how the research team used deep learning and artificial intelligence to identify tumor-infiltrating lymphocytes (TILs) maps, or patterns. Credit: Stony Brook University
By combining data on pathology images of 13 types of cancer and correlating that with clinical and genomic data, a Stony Brook University-led team of researchers are able to identify tumor-infiltrating lymphocytes (TILs), called TIL maps, which will enable cancer specialists to generate tumor-immune information from routinely gathered pathology slides. Published in Cell Reports , the paper details how TIL maps are related to the molecular characterization of tumors and patient survival. The method may provide a foundation on how to better diagnose and create a treatment plan for cancers that are responsive to immune-based anti-cancer therapy, such as melanoma, lung, bladder, and certain types of colon cancer.
The gold standard for cancer diagnosis remains the pathology report from a biopsied tumor tissue. Diagnosis plays a leading role in how a patient will be treated. In certain situations and with forms of cancer treated with immune-based therapies, pathologists are also tasked with making observations on the immunologic features of the tumor tissue to determine which patients are most likely to benefit from these therapies. TILs are unleashed by immunotherapies to destroy cancer cells.
"This paper demonstrates that we can now use deep learning methods such as artificial intelligence to extract and classify patterns of immune cells in ubiquitously obtained pathology studies, and to relate immune cell patterns to the many other types of cancer patient molecular and clinical data," says Joel Saltz, MD, PhD, the Cherith Chair of Biomedical Informatics at Stony Brook University and lead author of the paper, titled "Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images."
The research includes researchers from Stony Brook University, the University of Texas MD Anderson Cancer Center, Emory University, and the Institute for Systems Biology. The work stems from the efforts of The Cancer Genome Atlas (TCGA) project.
In the study, the researchers applied machine learning to digitized pathology images to characterize patterns of immune infiltration present in 4,759 TCGA patients and within 13 cancer types processing more than 5,000 digital images from the cancer types to create a "computational stain" for each. With these, they created TIL maps as a potential new guide to diagnosis and treatment planning.
TCGA is a decade long comprehensive effort spearheaded by the National Cancer Institute (NCI) and the National Human Genome Research Institute in collaboration with the cancer research community worldwide. The TIL map study is part of a cohort of 27 manuscripts published in Cell Press communicating results by the TCGA PanCancer Atlas Initiative, which has compared and contrasted molecular features of all TCGA tumor samples from more than 10,000 cases comprising 33 different forms of cancer.
"Developing machine learning tools such as this proof of principle project to map lymphocytic infiltration patterns is important for research reproducibility in immune-oncology and will also allow these approaches to begin to be deployed as decision support for pathologists as we evaluate and report our cases for routine decision-making," says Alexander Lazar, MD, PhD, Professor of Pathology & Genomic Medicine at MD Anderson and a co-author.
The investigators were able to develop the method and proof of concept with the assistance of data collection and calculations by way of high-performance computing systems available through Stony Brook's Institute for Advanced Computational Science and Division of Information Technology.
More information: Joel Saltz et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images, Cell Reports (2018). DOI: 10.1016/j.celrep.2018.03.086
Journal information: Cell Reports
Provided by Stony Brook University