Moving a cardiac regulatory protein to the right place
The largest known protein, titin, named for the Titan giants of Greek mythology, is a molecular spring found in cardiac muscle. It provides structural support and maintains tension during muscle stretching. Mutations in the titin gene, and in the gene encoding its regulatory protein RBM20, cause dilated cardiomyopathy (DCM), in which the left ventricle of the heart is enlarged and weakened, and cannot pump blood around the body efficiently.
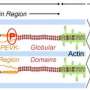
The RBM20 protein is divided into domains that have remained unchanged throughout evolution, thus sharing identical sequences among different species. Such regions are usually functionally important, but the role of the fully conserved 5-residue RSRSP stretch within an RS-rich region, full of arginine (R) and serine (S) amino acids, had not been investigated. However, this stretch is the site of many different mutations in DCM patients, a mutation "hotspot," so it was ripe for exploration.
An international team of researchers led by Tokyo Medical and Dental University (TMDU) investigated the RSRSP stretch and showed it to be essential for nuclear localization of the RBM20 protein. Chemical modification of part of this stretch was also found to be important in the role of RBM20 as a titin regulator. The study was reported in Scientific Reports.
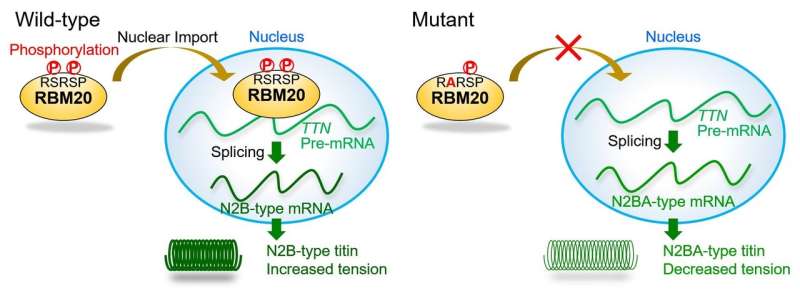
RBM20 controls a step in gene expression of the titin gene known as splicing, whereby introns are removed to leave the protein-coding exons behind as messenger RNA. Intron-exon processing can exclude certain exons, resulting in two major versions of titin that have distinct effects on normal cardiac function as well as cardiac dysfunction leading to DCM progression.
"We found mutations in DCM patients and examined the effect of mutations, especially those in the RSRSP stretch of RBM20," study authors Hidehito Kuroyanagi and Akinori Kimura say. "Wild-type RBM20 was localized in the nucleus where splicing occurs, whereas the mutations in the RSRSP stretch prevented the protein from localizing there and carrying out its function." However, the mutations had no effect on the actual role of RBM20 in splicing per se.
Mice genetically engineered to carry an RBM20 mutation of the RSRSP stretch seen in DCM patients were unable to express the smaller version of titin like the patients because of the effect of the mutation on the RBM20 function as a splicing regulator. These mice will prove useful as a DCM disease model for future in vivo work.
The team then showed that amino acids in the RSRSP stretch undergo phosphorylation, which is the enzyme-controlled addition of a phosphate group that is often involved in protein activation. "For the first time, we found that two serine residues in the RSRSP stretch are constitutively phosphorylated," corresponding author Hidehito Kuroyanagi explains. "Moreover, the simultaneous phosphorylation of both serine residues was essential for the nuclear localization of RBM20."
More information: Rie Murayama et al, Phosphorylation of the RSRSP stretch is critical for splicing regulation by RNA-Binding Motif Protein 20 (RBM20) through nuclear localization, Scientific Reports (2018). DOI: 10.1038/s41598-018-26624-w