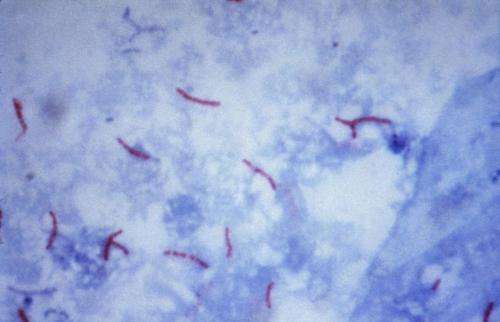
This photomicrograph reveals Mycobacterium tuberculosis bacteria using acid-fast Ziehl-Neelsen stain; Magnified 1000 X. The acid-fast stains depend on the ability of mycobacteria to retain dye when treated with mineral acid or an acid-alcohol solution such as the Ziehl-Neelsen, or the Kinyoun stains that are carbolfuchsin methods specific for M. tuberculosis. Credit: public domain
A team of researchers from the University of Wisconsin, the University of Iowa and the Norwegian Institute of Public Health has carried out genetic studies of tuberculosis to learn more about its lineage and to compare it with transmission pathways through early human migration. They have outlined their study and results in a paper available on the bioRxiv preprint server. Team lead Caitlin Pepperell also presented the group's findings at this year's ASM Microbe 2018 meeting.
The lineage of TB is unique, the team reports, because it can only be transmitted by humans. It was for that reason that they set out to study the bacterium that causes it as it has evolved over time. But they also wanted to tie such possible changes to human history or historical events.
To learn more about the history of the bacteria, the team collected 552 samples of Mycobacterium tuberculosis genomes and sequenced them. They then used the data to build a phylogenetic tree, which they mapped to major human migrations.
The team found that mass dispersal of the bacteria coincided with major human migration routes, such as the Silk Road. They note that the spread was both efficient and complex regarding the migration patterns. They report being able to trace exchanges between people traveling in eastern Africa and Southeast Asia, with those that were living there. They noted also a definite lack of dispersal out of China and some other parts of Asia, which they attributed to isolationism.
As part of their effort, the team reported that they studied over 1,000 TB isolates, paying particular attention to those that are known to be drug resistant. They also included some notable adaptations humans have developed in response to infections. They note, for example, that sanatoriums were built during the 1800s to house patients, keeping them away from others as they were treated. But they were surprised to find that the length of stay for such patients tended to increase over time, and that the numbers of people admitted grew. They also saw increases in the number of repeat patients.
More information: Mary B. O'Neill et al. Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia, bioRxiv (2017). DOI: 10.1101/210161
Abstract
Mycobacterium tuberculosis (M.tb) is a globally distributed, obligate pathogen of humans that can be divided into seven clearly defined lineages. How the ancestral clone of M.tb spread and differentiated is important for identifying the ecological drivers of the current pandemic. We reconstructed M.tb migration in Africa and Eurasia, and investigated lineage specific patterns of spread. Applying evolutionary rates inferred with ancient M.tb genome calibration, we link M.tb dispersal to historical phenomena that altered patterns of connectivity throughout Africa and Eurasia: trans-Indian Ocean trade in spices and other goods, the Silk Road and its predecessors, the expansion of the Roman Empire and the European Age of Exploration. We find that Eastern Africa and Southeast Asia have been critical in the dispersal of M.tb. Our results reveal complex relationships between spatial dispersal and expansion of M.tb populations, and delineate the independent evolutionary trajectories of bacterial sub-populations underlying the current pandemic.
© 2018 Medical Xpress