Lung cancer is the leading cause of cancer death among men and women. About 85 percent of lung cancers are non-small cell lung cancer. For a handful of these patients, therapies that target specific genetic mutations are effective. But for the majority of non-small cell lung cancer patients, targeted therapies are limited and many patients develop resistance to treatment, highlighting the need for other options.
Moffitt Cancer Center researchers are combining the large-scale study of proteins (proteomics) with a new data integration method to identify a previously unknown mechanism for midostaurin in lung cancer. Midostaurin is a drug approved by the United States Food and Drug Administration for the treatment of acute myeloid leukemia and advanced systemic mastocytosis. Their study was published in the journal Molecular & Cellular Proteomics.
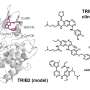
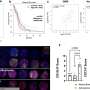
The research team led by Uwe Rix, Ph.D., associate member of the Drug Discovery Department at Moffitt, worked to identify all of the proteins that interact with midostaurin in non-small cell lung cancer cells in the lab. They then used a data analysis technique developed in the Rix lab to further examine the pathways associated with those proteins. In the end, the researchers identified three protein targets of midostaurin, TBK1, PDPK1 and AURKA, previously unknown to be important for midostaurin's mechanism of action in lung cancer cells.
This discovery allowed the team to design a combination therapy using midostaurin and BI2536, a protein inhibitor currently being investigated for the treatment of multiple cancers, which had a much greater effect on reducing non-small cell lung cancer cell growth than using either drug alone.
"Our integrated proteomics approach was particularly significant in the discovery of midostaurin's new mechanism of action, as none of the identified proteins are mutated at the gene level and would have been missed by traditional genomic screens," said Rix. "Utilizing protein pathway analysis in combination with functional proteomic techniques opens up the possibility for the identification of previously unknown actionable drug targets and combination therapies for many different cancers."
More information: Claudia Ctortecka et al, Functional proteomics and deep network interrogation reveal a complex mechanism of action of midostaurin in lung cancer cells, Molecular & Cellular Proteomics (2018). DOI: 10.1074/mcp.RA118.000713
Journal information: Molecular & Cellular Proteomics