Paradigm shift in tuberculosis treatment: Genome sequencing replaces standard resistance testing
An international research team from the CRyPTIC Consortium has taken a critical step forward in the fight against tuberculosis: a large-scale genome analysis of over 10,000 pathogen strains has shown that genome sequencing can improve tuberculosis treatment. Furthermore, the method has the potential to replace time-intensive phenotypic resistance testing. The results of this study, led by the University of Oxford, have been published in the current issue of the New England Journal of Medicine.
Tuberculosis (TB) is the deadliest infectious disease worldwide. According to a current report from the World Health Organisation (WHO), approximately 10 million people contracted TB and 1.6 million people died of the consequences of infection last year.
A major problem in tuberculosis treatment is the increased occurrence of multidrug-resistant strains (MDR-TB) that are no longer sensitive to commonly used drugs. A timely diagnosis of resistance is critical to treatment success. Classical resistance testing involves culturing bacteria in mediums containing antibiotics, which currently takes several weeks. Additionally, these tests are not used in many countries with high rates of TB. Consequently, many patients take the wrong drug combinations and subsequently have lower chances of being cured and of survival. More rapid and practical methods for resistance testing are urgently needed.
The CRyPTIC study examined over 10,000 bacterial genomes from 16 countries. The Research Center Borstel (FZB), Leibniz Lung Center, has donated data from over 600 TB strains obtained in Germany, Africa and Central Asia and was involved in evaluating the data.
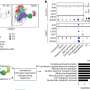
The study, which was also presented at the U.N. General Assembly's High-Level Meeting on Ending TB on 26 September 2018 in New York, shows that changes in pathogen genomes can be used to predict resistance to standard first-line drugs very accurately. Genome sequencing can precisely and individually determine the required drug combinations and save resources used in standard resistance testing. The FZB researchers are also working on establishing genome analysis in reference laboratories in central Asia and Africa.
"The study data enables new concepts for TB diagnostics to be developed. Additionally, standard resistance testing for commonly used drugs could soon be replaced by genome-based methods. This is a milestone and a paradigm shift in the fight against MDR-TB," says Stefan Niemann, head of the CRyPTIC study at the FZB and coordinator of the TB research field at the German Center for Infection Research (DZIF).
In England, the Netherlands and at the Wadsworth Center for Public Health in New York, the results have already led to a decrease in the use of bacterial cultures for resistance testing and a start to treatment that is solely based on genome sequencing results. In Germany, planning for a pilot study to evaluate the nationwide suitability of genome sequencing for diagnostics and surveillance is also currently underway at the Robert Koch Institute and the FZB.
More information: , Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing, New England Journal of Medicine (2018). DOI: 10.1056/NEJMoa1800474