Dr Jana Gerstmeier und Markus Werner investigate the effect of active compounds on the formation of endogenous signalling substances in immune cells during an inflammatory reaction. This will help to develop new active drugs with fewer side effects. Credit: Jan-Peter Kasper/FSU
An international research team led by Friedrich Schiller University in Jena has developed a highly sensitive cell model to study the complex effects – and side effects – of anti-inflammatory drugs, with the ultimate aim of preventing chronic inflammation.
Drugs such as ibuprofen or aspirin that relieve pain and lower fever are among the most frequently used drugs worldwide. They are used above all for the treatment of inflammatory reactions. However, in spite of their indisputable effectiveness and frequent use, we do not yet fully understand the underlying mechanisms of these drugs. In addition, when these drugs are taken, serious side effects occur and again, the causes of which have also not been sufficiently clarified.
The research team led by pharmacists Dr. Jana Gerstmeier and Prof. Oliver Werz of the University of Jena has developed a cell model with which they can find answers to these questions. As the team's scientists report in the specialist publication, The FASEB Journal, they have succeeded in clarifying the complex effect of active compounds that are administered on the formation of endogenous signalling substances in immune cells during an inflammatory reaction. In the future, this will make it possible to develop new active drugs with fewer side effects. Working groups from Harvard Medical School in Boston and the Karolinska Institute in Stockholm were also involved in the research work.
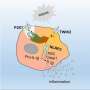
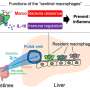
"Inflammation proceeds – roughly speaking – in two successive phases," explains Markus Werner, a doctoral student at the Chair of Pharmaceutical and Medical Chemistry of Jena University and first author of the study. During the initial phase, type 'M1' immune cells (macrophages) are active. They produce inflammatory messenger substances (prostaglandins and leukotrienes) from unsaturated fatty acids, which trigger typical symptoms such as fever and pain. After a few days, the second phase begins, in which the inflammation is resolved. In this phase, type 'M2' macrophages are active, which produce inflammation-resolving messenger substances from the fatty acids (called resolvins).
"Conventional drugs intervene equally in both phases," says Dr. Jana Gerstmeier. "They reduce the production of both proinflammatory messenger substances and inflammation-resolving mediators." This alleviates the first acute inflammatory reaction, but at the same time it also hampers the second phase in which the inflammation resolves. "There is a risk of inflammation not being stopped and continuing to progress, so that secondary diseases occur." Ideally, drugs should therefore reduce only the acute phase, but not impair the phase in which the inflammation resolves.
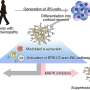
The newly developed cell model enables researchers to investigate the efficacy of drugs in both inflammatory phases. "For this purpose, we use human immune cells (M1 and M2), which we pre-treat with the drug to be tested before inducing an inflammatory reaction using pathogenic bacteria," explains Jana Gerstmeier. The messenger substances released by the cells are analysed.
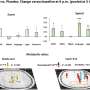
The special feature of the methodology developed in Jena is its sensitivity: The inflammation-resolving substances of the second phase are effective in concentrations about 1,000 times lower than the inflammatory signal substances of the first inflammatory phase. Very sensitive analytics are required in order to be able to detect these substances, and the Jena laboratory is one of the few laboratories in the world to have mastered this methodology. Using a mass spectrometer, several dozen mediator molecules that have been released are detected and an individual spectrum is created for each active ingredient. This allows conclusions to be drawn about the influence of the drug on the entire inflammatory process.
More information: Markus Werner et al. Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome, The FASEB Journal (2019). DOI: 10.1096/fj.201802509R
Journal information: FASEB Journal
Provided by Friedrich Schiller University of Jena