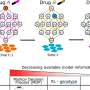
A new computational approach has been developed to reduce variability in common research biomarker tests, a promising step in improving the ability of biomedical researchers and basic scientists to reproduce data and facilitate more consistent results across laboratories and long-term projects. Researchers from Boston Medical Center (BMC) and Boston University School of Medicine (BUSM), developed a new software program, called ELISAtools, which provides a stable platform to compare data from research-use-only assay kits and minimize variability over months or even years. The results were published online in PLOS One.
Enzyme-Linked Immunosorbent Assay (ELISA) tests are used globally across clinical, biomedical and basic research fields to measure biomarkers in a range of mediums including blood, plasma, and urine. Clinical ELISA test kits used in the hospital setting are regulated to ensure tight quality control boundaries for accuracy and consistency. However, the hundreds of commercially available research-use-only ELISA test kits are not regulated, which often leads to noticeable variability in results over time, between testing kits, and across different laboratories.
The BMC-BUSM research team unexpectedly encountered high variability from one ELISA test kit during a project for the National Cancer Institute measuring thrombosis and inflammation biomarkers in the plasma of cancer subjects and healthy donors. After the first year of the project, they realized the data was changing significantly as they received different shipments of the kit from the manufacturer. After a thorough examination, they determined differences in the ELISA kit was causing the issue. They had research data from over 400 patient samples that could not be compared due to these differences in the ELISA kits. To solve this problem, the research team created the ELISAtools software program to reduce future variability in test results.
"After implementing this software, the variability in test results dropped from over 60 percent, to under 9 percent, well within our quality control limits," says Deborah J. Stearns-Kurosawa, Ph.D., senior author of the study and associate professor of pathology and laboratory medicine at BUSM. "We work on studies that go on for years, and this tool creates a constant, level playing field that we believe will improve accuracy and clinical utility of research."
More information: Feng Feng et al, A computational solution to improve biomarker reproducibility during long-term projects, PLOS ONE (2019). DOI: 10.1371/journal.pone.0209060
Journal information: PLoS ONE
Provided by Boston Medical Center