Credit: CC0 Public Domain
A key mechanism controlling tissue structure, which could help identify drugs that make it harder for cancer cells to spread, has been identified by researchers at the Francis Crick Institute.
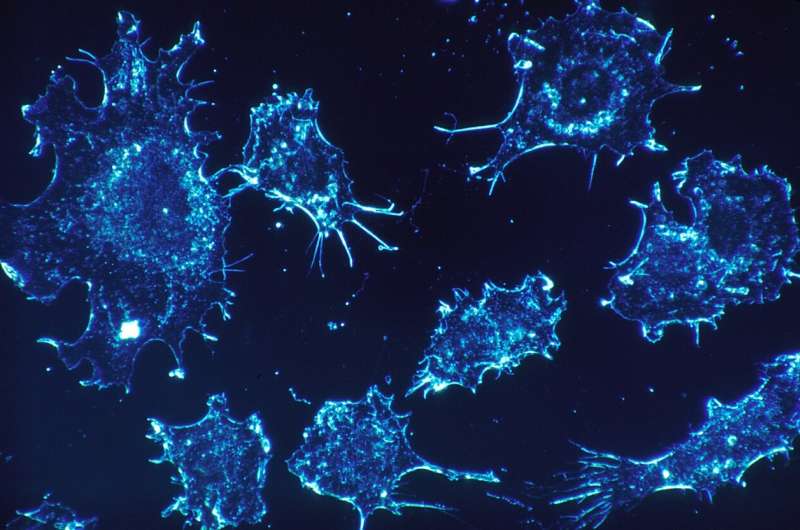
The two studies, published in Nature Materials and PLOS Computational Biology, explain the mechanism that causes changes in the structure of tissues. Using experimental and computational biology, the studies identified how collisions between cells help to create different tissue structures. Some of these structures help cancer spread and the authors also found drugs that inhibit this process.
All tissues in the body contain a scaffold of proteins that maintain their structure. This scaffold degrades as people age, leading to signs of aging such as wrinkles.
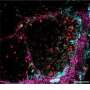
Cancerous cells can corrupt the shape of the scaffold to create a tissue structure with pathways that lead away from the tumour and into the surrounding tissue. The cancerous cells take advantage of these super-highways by moving along them to spread into new areas.
However, while it is known that these super-highways are linked to cancer progression, there is little understanding about the mechanisms that control the formation of these tissue structures.
"These super-highways provide roads for cancer cells to travel out of tumours and spread more widely in the tissue, having potentially disastrous consequences for the patient," says Danielle Park, research scientist in the Crick's Tumour Cell Biology Laboratory. "By understanding more about how this type of structure is formed, we can then look at finding ways to stop it and impose a road block on the spread of cancer cells."
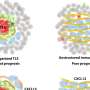
By combining insights from laboratory experiments with results from a new computer model which the team developed, the researchers discovered how a tissue scaffold's shape is affected by how it interacts with specialist cells which help to build the scaffold, known as fibroblasts.
"Our computer model draws inspiration from how birds and fish, the fibroblasts in our case, move closely together in flocks or schools, despite being in very large groups," says Esther Wershof, a Ph.D. student in the Biomolecular Modelling Laboratory at the Crick. "With this model, it's much easier to study the relationship between cells and the scaffold than if we could only watch it in real-life."
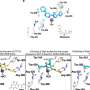
Further research found that a key factor in the formation of tissue with super-highways is how the fibroblasts act when they collide with each other. By identifying a protein (TFAP2C) that regulates these collisions, the researchers could then look for drugs that inhibit this protein and, in turn, disrupt the creation of super-highways and the spread of cancer cells. Using existing datasets and experiments in the laboratory, the study successfully identified five drugs that affected the formation of super-highways in the tissue.
"This work is a great example of insights that can be uncovered when experimental and computer biologists collaborate. By working together, using a range of techniques and bringing different expertise to the table, we can reach new understandings about how the body works and how we can better treat disease," said Paul Bates, group leader of the Biomolecular Modelling Laboratory at the Crick.
More information: Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions, Nature Materials (2019). DOI: 10.1038/s41563-019-0504-3 , nature.com/articles/s41563-019-0504-3
Journal information: Nature Materials , PLoS Computational Biology
Provided by The Francis Crick Institute