Coronavirus 2019-nCoV: The largest meta-analysis of the sequenced genomes of the virus

The largest analysis of coronavirus 2019-nCoV genomes that have been sequenced so far confirms that the virus originates in bats and shows a low virus heterogeneity. At the same time, researchers identified a hyper-variable genomic hotspot in the proteins of the virus responsible for the existence of two virus subtypes. The leading author of this study, published in the Journal of Medical Virology, is Federico M. Giorgi, bioinformatics researcher at the Department of Pharmacy and Biotechnology of the University of Bologna.
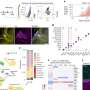
The data released by the World Health Organization reveal that to date, the coronavirus 2019-nCoV has infected 28,276 people, of whom 565 died. This new study analyzed the genomes of the 56 coronavirus strains sequenced in different parts of the world, including those extracted from the two Chinese patients held at the Infectious Disease Ward of Lazzaro Spallanzani Hospital in Rome, Italy. This is the most comprehensive study of coronavirus genomes so far conducted.
Researchers confirmed the notion that the virus probably originates from a zoonotic pathogen: its closest relative, which was isolated in the past few weeks, matches the coronavirus sequence EPI_ISL_402131 found in the Rhinolophus affinis, a medium-size Asian bat of the Yunnan Province (China). The human coronavirus genome shares at least 96.2% of its identity with its bat relative, while its similarity rate with the human strain of the SARS virus (severe acute respiratory syndrome) is much lower, only 80.3%.
The researchers have also discovered that all the existing DNA sequences of coronavirus are very similar, even if they come from different regions of China and from various parts of the world—the genomes obtained from patients since the beginning of the outbreak share a sequence identity over 99%. "The virus shows low heterogeneity and variability—this is good news," explains Federico M. Giorgi. "With a homogeneous viral population, potential drug therapies are deemed to be more effective on everyone."
However, the study identified for the first time a hyper-variable hotspot in the virus proteins, eventually pinpointing two virus subtypes. The latter differ only by a single amino acid, which is able to change the sequence and the structure of ORF8-encoded protein, a virus component yet to be characterized.
More information: Carmine Ceraolo et al. Genomic variance of the 2019‐nCoV coronavirus, Journal of Medical Virology (2020). DOI: 10.1002/jmv.25700