AI reveals differences in appearance of cancer tissue between racial populations
Scientists at Case Western Reserve University are using Artificial Intelligence (AI) to reveal apparent cellular distinctions between black and white cancer patients, while also exploring potential racial bias in the rapidly developing field of AI.
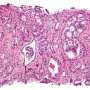
Their most recent published research asserts that AI analysis of digitized images of cancer tissues reveals critical variations between black and white male prostate cancer patients. The work also suggests the new population-specific information—in addition to image detail on tissue slides also analyzed by computers—could substantially improve care for black patients with prostate cancer.
"On one level, we're simply trying to understand and answer this question: "Are there biological differences in the disease, in the cancer, that are a function of your ethnicity or your race?'" said Anant Madabhushi, the F. Alex Nason Professor II of Biomedical Engineering at Case Western Reserve and senior author on a study published today in Clinical Cancer Research, a journal of the American Association for Cancer Research. "In other words, is there something else going on that can't be explained by other disparities? The answer appears to be 'yes.'"
This new work on prostate cancer builds on mounting evidence that clear biological differences between races can be discovered at a cellular level in the analysis of cancer cells—information which can be useful to tailor medical care to specific groups and individuals within those populations.
Implicit in all of the ongoing research, Madabhushi said, is the larger question about whether the racial differences being discovered at the cellular level are revealing a research bias at the human level.
"Even as we do this groundbreaking research, we can't allow ourselves to get trapped into trusting these models blindly," he said, "so we need to question whether we are considering all populations (and) ask how diverse our research pool is."
Prostate cancer study
Racial differences were a key component in the most recent research work. The prostate cancer study was performed over three years at six sites and involved nearly 400 men with the disease.
One of the critical questions in management of prostate cancer patients is to identify which men following prostate surgery are at higher risk of disease recurrence and could benefit from adjuvant therapy.
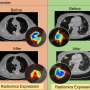
The patient pool in this study, however, was about 80% white and 18% black, "so the model was biased toward the majority population," Madabhushi said. "Once we found the variations, applying the model to all would be doing a disservice to that one population."
Once researchers created a race-specific model, the accuracy in determining which black patients would have a recurrence of the cancer increased six-fold, Madabhushi said.
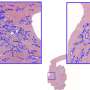
Like previous cancer research led by Madabhushi's lab, the scientists asked the computer to look for patterns not only from images of the tumor itself, but at tissue outside the tumor, known as the stroma.
In doing so—in this and other cancer studies—they have been able to successfully tell, among other things, which patients would respond well to chemotherapy, immunotherapy or even, in some cases, whether cancer would return or how long a patient might live.
Aside from non-melanoma skin cancer, prostate cancer is the most common cancer among men in the United States and one of the leading causes of cancer death among men of all races, according to the Centers for Disease Control.
Further, while surgical resection of the prostate—known as a radical prostatectomy—is performed for about 75,000 newly diagnosed patients each year, 30% to 40% will see the cancer return, Madabhushi said.
In this case, armed with the knowledge of which patients had a recurrence of the cancer, scientists were able to retroactively see visual signals in tissue slides from their initial diagnosis to determine which patients would suffer that recurrence.
Lead authors who collaborated with Madabhushi on the paper included Hersh Bhargava, a Ph.D. student at the University of California-San Francisco and Patrick Leo, a graduate student of biomedical engineering at Case Western Reserve University.
Bhargava said the researchers were able to "look at, and actually measure, hundreds of thousands, even millions, of cancer cells to see features that a human could never see—including structural characteristics."
"It's clear from the existing scientific literature that there are racial disparities in all cancers, but it appears that especially in prostate cancer that those differences can't be explained by access to care or socioeconomic status—but rather that there is a biological component to how the cancers manifest differently between black and white patients."
Leo said that the research was focused on a population-disparities element that has not been recognized until now in what he called the "AI-for-health space."
"We know now that the risk is that if you just build a model for all patients, you will actually perform worse for patients in the minority and that's something we cannot accept, even if it's not something we did intentionally. So, if you want a model to work on patients from all populations, you have to deliberately include a population-specific aspect."
More information: Hersh K. Bhargava et al. Computationally Derived Image Signature of Stromal Morphology Is Prognostic of Prostate Cancer Recurrence Following Prostatectomy in African American Patients, Clinical Cancer Research (2020). DOI: 10.1158/1078-0432.CCR-19-2659