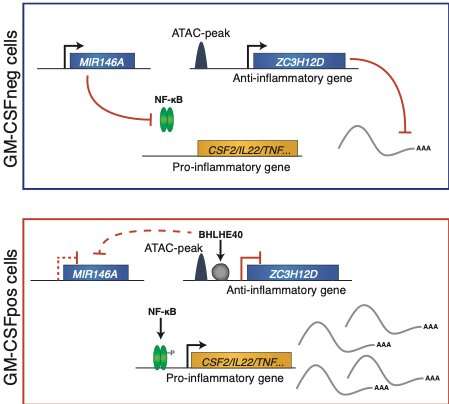
Schematic diagram describing the regulatory module identified in this study. Using GM-CSF secretion to enrich inflammatory lymphocytes, the authors identified a regulatory module that influences the activity of these proinflammatory and potentially pathogenic cells. In GM-CSFnegative cells, mechanisms are in place to actively repress the expression of inflammatory mediators, including high levels of miR-146a expression (a negative regulator of NF-B activation), and relatively higher expression of ZC3H12D, an RNase enzyme involved in the negative regulation of cytokine expression. Conversely, in GM-CSFpositive cells, the expression of the transcriptional repressor BHLHE40 leads to the direct downregulation of ZC3H12D expression, and indirectly affects also miR-146a. This in turn allows full-blown activation and expression of inflammatory cytokines. Credit: Università della Svizzera italiana
A research team led by the Institute for Research in Biomedicine (IRB) in Bellinzona, Switzerland, affiliated with Università della Svizzera italiana (USI), has identified a molecular network required to balance the responses of human lymphocytes. The activities of these immune cells must be carefully regulated in order to ensure effective defense of the organism. At the same time, the risk of damaging healthy tissues must be limited. Tissue damage may derive from excessive, unrestrained immune responses that cause many chronic inflammatory diseases such as multiple sclerosis. With this work, the IRB researchers have identified a molecular module involved in regulating the features that characterize highly proinflammatory and potentially pathogenic lymphocytes.
This study, published in the scientific journal Nature Immunology, was carried out in the laboratory of Silvia Monticelli at IRB, in collaboration with the laboratory of Gioacchino Natoli at the European Institute of Oncology (IEO) in Milan (Italy), and it was funded primarily by the Swiss National Science Foundation, the Swiss National Center for Competence in Research (NCCR) 'RNA & Disease', the Swiss Multiple Sclerosis Society and the Ceresio Foundation.
Background
Within the immune system, T helper lymphocytes are essential to orchestrate protective responses against invading pathogens, but they can also lead to immune-mediated chronic disorders by mounting responses against normal, healthy tissues. Because these cells are present in both healthy and diseased individuals, mechanisms must exist to control their pathogenic activation.
The discovery
In this study, the researchers identified a network of molecular regulators able to favor or repress a proinflammatory phenotype in T lymphocytes. In particular, the transcriptional regulator BHLHE40 acted as an enforcer of a highly inflammatory program by repressing the expression of negative regulators of inflammation, including microRNAs and RNA-binding proteins. Overall, this work advances our knowledge on the regulation of inflammatory programs and responses in human T lymphocytes, with the potential to contribute to our understanding of chronic inflammatory diseases.
Stefan Emming, a co-author of the article, says, "About five years ago, we set off trying to understand what makes a T lymphocyte pro-inflammatory and potentially pathogenic. After a great team effort, we identified a molecular network that can tip the balance, highlighting the importance of fine tuning T cell responses to avoid chronic inflammation and autoimmune diseases."
Niccolò Bianchi, another co-author, says, "In this work, we unraveled a molecular network regulating the production of proinflammatory cytokines in T cells, identifying BHLHE40 as the leading factor. This important discovery broadens our horizon in understanding chronic inflammatory diseases."
Co-author Sara Polletti says, "I am very proud to have been involved in this collaborative multidisciplinary project that led to very important results with implications in various aspects of immunity, possibly including cancer immunotherapy."
More information: Stefan Emming et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes, Nature Immunology (2020). DOI: 10.1038/s41590-020-0622-8
Journal information: Nature Immunology
Provided by Università della Svizzera italiana