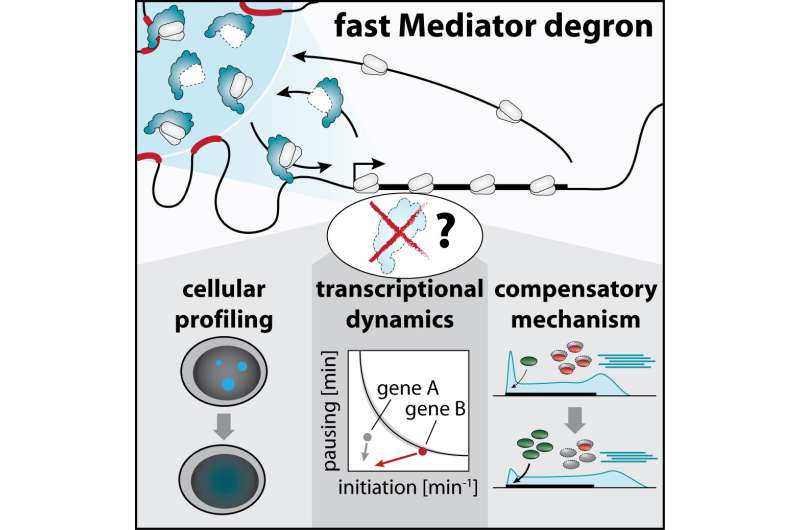
Visualizing gene expression dynamics revealed how Mediator directs the delivery of the cellular gene-copying machine (RNA polymerase II) particularly to a subset of cell identity genes. Credit: Martin Jäger / CeMM
Using targeted protein degradation technology, researchers at CeMM, the Research Center for Molecular Medicine of the Austrian Academy of Sciences, set out to understand the primary role of a key regulator of transcription, the human Mediator complex. Mediator, a 30-subunit molecular machine, was believed to be a factor that is generally required for the transcription of all protein-coding genes in human cells. However, the recent study by Georg Winter's group at CeMM discovered that Mediator particularly safeguards the transcription of a small set of cell-type-specifying genes. In collaboration with Patrick Cramer's lab at the Max Planck Institute for Biophysical Chemistry in Göttingen, the scientists revealed mechanistic details of how Mediator enables cells to robustly exert their dedicated functions. The study, "Selective Mediator dependence of cell-type-specifying transcription," was published in Nature Genetics, on 1 June 2020.
Georg Winter's lab at CeMM uses chemical tools to understand the molecular basis of gene control and aberrations thereof in cancer. To do so, the team combines particularly fast-acting protein ablation systems ("degradation Tag"/ "dTAG", co-developed by Georg Winter in Jay Bradner's lab at the Dana Farber Cancer Institute, in Boston) with precise and unbiased measurements of gene activity at high kinetic resolution. Together with Patrick Cramer's lab and others, the international team of scientists now unveiled how the Mediator molecular machine mechanistically directs gene activation, which had long remained poorly understood owing to challenges in experimental manipulation.
Mediator was identified over 30 years ago as a molecular bridge that allows DNA-binding proteins called transcription factors to communicate with the cellular gene-copying machine RNA polymerase II (Pol II), to activate target genes. Understanding the direct role of Mediator in this process requires technologies that rapidly block Mediator function and measure changes in Pol II activity within the subsequent minutes. Using targeted protein degradation, scientists in the Winter lab now managed to rapidly remove individual parts of the Mediator complex to determine whether the copying of all human genes depends to the same extent on Mediator integrity. In contrast to the existing paradigm, the study first-authored by CeMM Ph.D. student Martin Jaeger suggests that Mediator is not generally required for transcription of all genes. Rather, the data imply that Mediator selectively safeguards the expression of a small set of genes, which form densely connected regulatory circuits to instruct cell-type-specific functions.
By visualizing Pol II enzymes in cells, the scientists observed that Mediator nucleates large clusters of transcription machinery components that are thought to form around regulatory DNA regions called super-enhancers. These super-enhancers direct the expression of cell-type-specific genes by physically touching their target genes, which are often millions of DNA bases away. When Mediator was degraded, the large Pol II clusters rapidly disappeared, but super-enhancers still seemed to touch their target genes, indicating that Mediator was not required to maintain these DNA contacts.
Together with experts from the Cramer lab in Goettingen, however, the team observed dramatic changes in the dynamics of Pol II turnover at super-enhancer-driven genes after Mediator loss. The data suggested that Mediator clusters drove highly efficient recruitment of Pol II enzymes to these cell-type-specific genes, allowing their copying at remarkable efficiency. At the same time, the majority of other genes seemed to be surprisingly mildly affected by acute Mediator loss, which made the scientists suspect that they undergo some unexpected type of compensatory boost.
The team looked for this compensatory mechanism, and found that promoter-proximal pausing of Pol II was globally reduced in response to Mediator loss. Pausing is a phenomenon whereby Pol II enzymes are held back for several minutes right after they start copying a gene. The waiting Pol II constitutes a roadblock that prevents the passing of other Pol II enzymes behind it, thus limiting the number of RNA copies produced in a given timespan. After Mediator degradation, the scientists observed that the main factor responsible for signaling pause release, called cyclin dependent kinase 9 (CDK9), more efficiently bound to DNA in cells and more actively tagged its protein targets with a phosphate group. Chemically blocking CDK9 activity made the transcriptional defects of Mediator ablation less selective for super-enhancer target genes, which highlights that the uncovered CDK9 activation shapes how cells react to acute Mediator loss. This unexpected finding suggested that pausing may have evolved as a buffering capacity to rapidly react to and partially compensate acute defects in Pol II recruitment.
The study opens up new avenues to understanding the composition of Mediator-nucleated Pol II clusters and how CDK9 is so efficiently activated in response to transcriptional stresses. Furthermore, Georg Winter imagines future possibilities to perhaps tackle diseases where Mediator function went awry: "Very preliminarily, our work might also have revealed some potential for Mediator as a drug target, and next steps might be to develop direct degrader molecules against this complex."
More information: Martin G. Jaeger et al, Selective Mediator dependence of cell-type-specifying transcription, Nature Genetics (2020). DOI: 10.1038/s41588-020-0635-0
Journal information: Nature Genetics
Provided by CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences