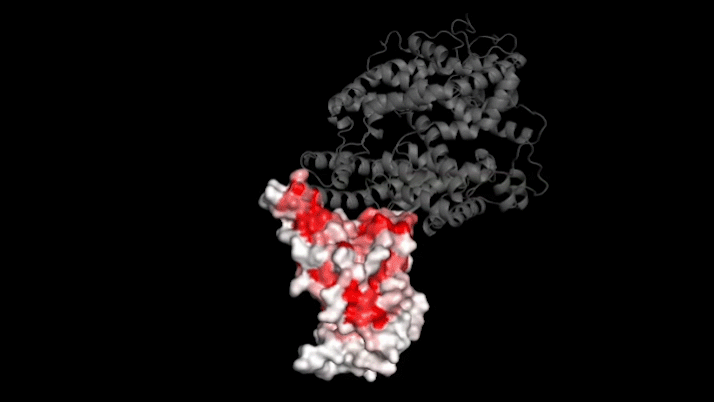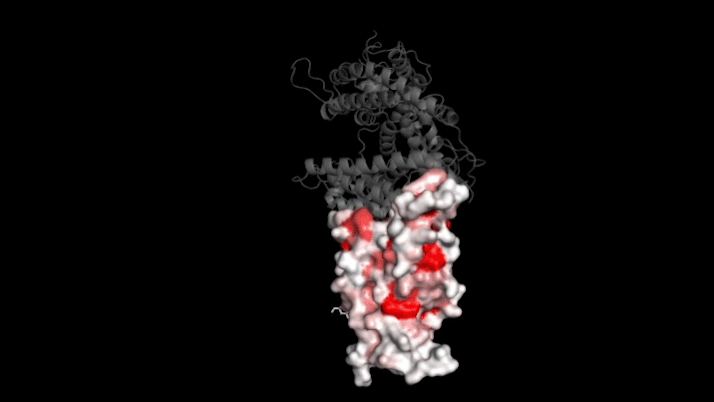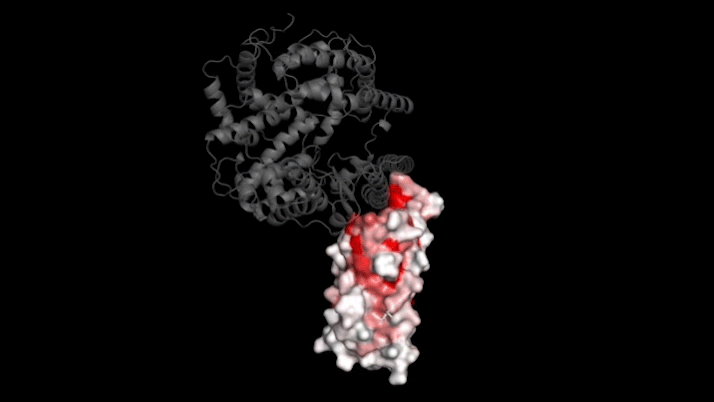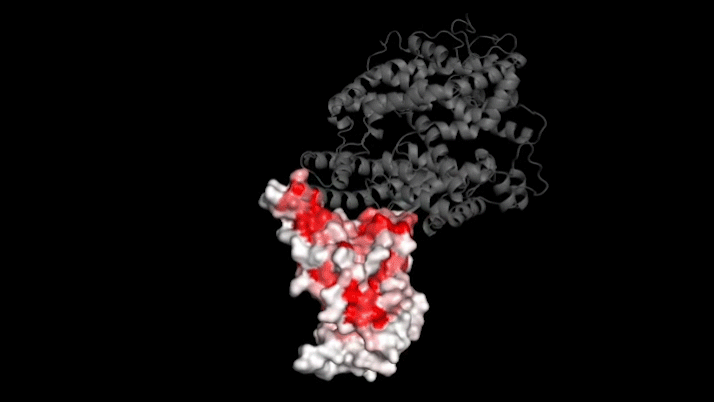
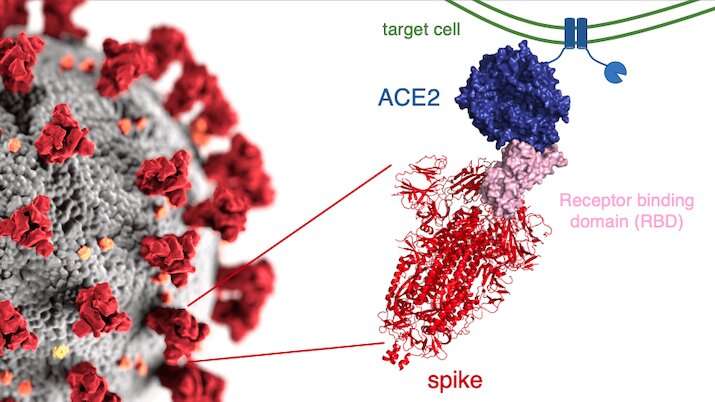
SARS-CoV-2 infects human cells by binding to a protein called ACE2 (grey). In this 3D model, dark red represents parts of the virus’s receptor binding domain where mutations greatly influence binding. Credit: Tyler Starr/Bloom Lab
As the novel coronavirus spreads, it's picking up new mutations—for better and for worse.
Now, Howard Hughes Medical Institute Investigator Jesse Bloom and his colleagues have cataloged how nearly 4,000 different mutations alter SARS-CoV-2's ability to bind to human cells.
Their data, publicly available online as an interactive map, is a new resource for researchers developing antiviral drugs and vaccines to fight COVID-19, the infectious disease caused by SARS-CoV-2. The work also reveals how individual mutations may affect the virus's behavior, the team reports August 11, 2020 in the journal Cell.
"We don't know how the virus will evolve, but now we have a way to look at the mutations that can occur and see their effects," says Bloom, a virologist at the Fred Hutchinson Cancer Research Center.
Each time a virus replicates, it can pick up new genetic mutations. Many of these mutations have no effect on a virus's behavior. Others could make the virus better or worse at infecting people. To what extent mutations might be making SARS-CoV-2 more dangerous has been an open—and controversial—question. Doctors and scientists have analyzed genetic differences in virus samples collected from COVID-19 patients around the world, hunting for clues to the disease's spread. But until now, no one had comprehensively linked potential mutations to their functional effect on SARS-CoV-2.
The new study focused on mutations to a key part of SARS-CoV-2—its "spike protein." This protein binds to a protein on human cells called ACE2, a necessary step for infection. Mutations in the spike protein could change how well SARS-CoV-2 sticks to—and thus infects—human cells.
Researchers are studying mutations to the part of SARS-CoV-2 (red and pink) that makes contact with the ACE2 protein (blue) on human cells. The work could reveal how the virus’s ability to infect cells may change over time. Credit: Tyler Starr/Bloom Lab and Alissa Eckert/MSMI; Dan Higgins/MAMS
Bloom's team bred yeast cells to display a fragment of the spike protein on their surface. This fragment, called the receptor binding domain, makes direct contact with ACE2. The researchers systematically created thousands of versions of the fragment—each with different mutations. Then they measured how well these mutated fragments stuck to ACE2. That let them assess how various mutations might affect the function of the binding domain.
The data show that many possible mutations could make the virus bind to human cells more strongly. But those mutations don't seem to be gaining a foothold in circulating versions of the virus.
"This would suggest that there's some sort of sweet spot, where if the virus can bind ACE2 pretty well, then it's able to infect humans," Bloom says. "Maybe there's no evolutionary need for it to get better."
Other mutations made it harder for the spike protein to bind to cells or prevented the protein from properly folding into its final shape, the team found. Versions of the virus with these mutations might be less likely to gain a foothold because they can't infect cells as effectively. The team's targeted lab tests aren't a perfect proxy for how mutations will affect the virus in the wild, where many other factors influence how effectively it can spread—but they're a useful starting place.
The data will also be valuable for researchers designing drugs and vaccines to fight COVID-19, says Tyler Starr, a postdoc in Bloom's lab who led the project alongside graduate student Allie Greaney. Understanding the consequences of different mutations can guide the development of drugs that will continue to work as the virus changes over time. Plus, Starr says, "it's becoming clear that antibodies that stick to this part of the virus are really good, protective antibodies that we would want to elicit with a vaccine."
Study coauthor Neil King and his lab at the University of Washington is already working on such vaccines. His team is designing artificial proteins that mimic components of the virus. As part of a vaccine, such proteins could potentially train people's immune systems to produce antibodies that target the coronavirus. The researchers modify the artificial proteins to make them more stable and easier to produce in large quantities than the natural versions of the proteins.
The data from Bloom's team offers a roadmap to making those modifications. "Normally, when we're trying to figure out how to make a protein better, we're shooting in the dark," says Daniel Ellis, a graduate student in King's lab. "The information they've given us is kind of like a cheat sheet. It makes our lives amazingly easier."
More information: Tyler N. Starr et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding, Cell (2020). DOI: 10.1016/j.cell.2020.08.012
Journal information: Cell
Provided by Howard Hughes Medical Institute