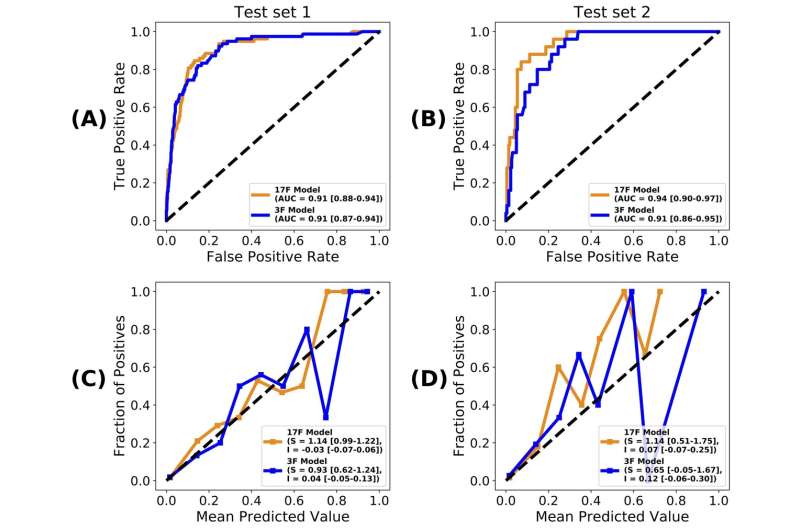
Evaluation results for test datasets 1 (A) and 2 (B) are shown here in terms of the ROC curves obtained, as well as their AUC scores, with 95% CIs in parentheses. Calibration curves of the 3F and 17F models on test datasets 1 (C) and 2 (D), with the slopes and intercepts of all the curves, along with their 95% CIs in parentheses. AUC=area underthe ROC curve. ROC=receiver operating characteristic. Credit: Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai
Given the toll that the COVID-19 pandemic has taken on people's health and lives worldwide, it is crucial to be able to accurately predict patients' outcomes, including their chances of mortality from the disease. Using the largest clinical dataset to date, and a systematical machine learning framework, the research team at Mount Sinai identified an accurate and parsimonious prediction model of COVID-19 mortality.
This model was based on only three routinely collected clinical features, namely patient's age, minimum oxygen saturation over the course of their medical encounter, and type of patient encounter (inpatient vs outpatient and telehealth visits).
This model could yield an additional "vital sign" that is assessed regularly during a patient's hospital course, that can be integrated into the clinical care flow of a COVID-19 patient. Clinical teams could use results from the prediction model throughout COVID-19 patients' hospital courses to flag individuals at high risk of death so that they can promptly focus treatment and attention on such individuals to prevent their mortality.
Using the largest development dataset yet (n=3841), and a systematic machine learning framework, we developed a COVID-19 mortality prediction model that showed high accuracy (AUC=0·91) when applied to test datasets of retrospective (n=961) and prospective (n=249) patients. This model was based on three clinical features: patient's age, minimum oxygen saturation over the course of their medical encounter, and type of patient encounter (inpatient vs outpatient and telehealth visits).
"Predicting mortality among patients with COVID-19 who present with a spectrum of complications is very difficult, hindering the prognostication and management of the disease," said Dr. Gaurav Pandey, Assistant Professor of Genetics and Genomic Sciences. "We aimed to develop an accurate prediction model of COVID-19 mortality using unbiased computational methods, and identify the clinical features most predictive of this outcome."
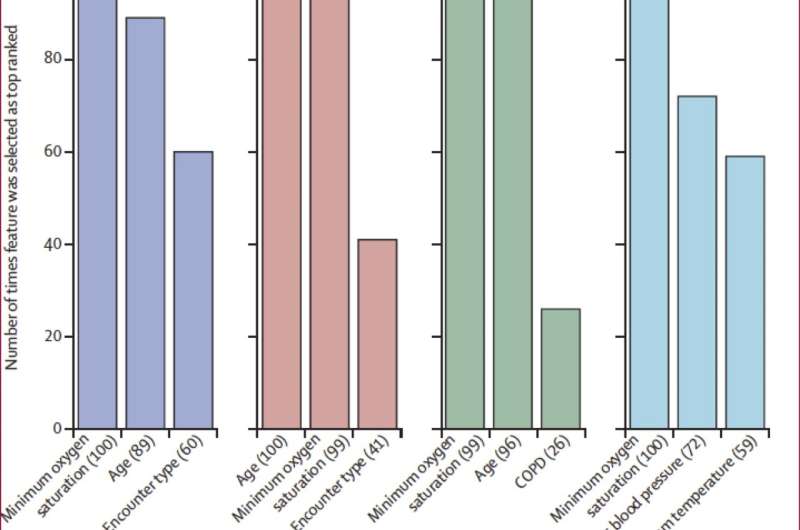
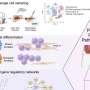
Top predictive features selected for the four classification algorithms (A) Top three predictive features identified using the recursive feature elimination method for the four classification algorithms across the 100 runs used to select the most discriminative features and train the corresponding candidate prediction models; the values in parentheses indicate the number of times the feature was selected as top ranked in the development dataset. Minimum oxygen saturation (B) and age (C) features, which were selected as top predictive features for all the four algorithms, are presented as violin plots showing the distributions of the values in the development dataset. In panels B and C, the black boxplots in the middle show the distribution of the values on the y axis, with the white dot indicating the median value; the width of the grey shape at a given value on the y axis indicates the probability of occurrence of that value in the population shown. The plots in panel B show that the median value (79%) of minimum oxygen saturation for the deceased group was significantly lower Credit: Icahn School of Medicine at Mount Sinai
More information: Arjun S Yadaw et al, Clinical features of COVID-19 mortality: development and validation of a clinical prediction model, The Lancet Digital Health (2020). DOI: 10.1016/S2589-7500(20)30217-X
Provided by The Mount Sinai Hospital