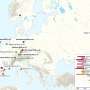
17th century skeletons of plague victims from the St. Gertrude cemetery in Riga, Latvia. From the individual marked with the number 701, scientists were able to reconstruct an entire plague genome. Credit: Guntis Gerhards
How do epedemics come to a stop? Scientists consider genetic mutation of the pathogens as a possible cause.
The plague killed about a quarter of the European population in the 14th century. How did it come to a stop? An international team of researchers led by Ben Krause-Kyora from the Institute of Clinical Molecular Biology (IKMB) at Kiel University in Germany (CAU) has now uncovered a mechanism that, among other factors, may have contributed to the end of the second pandemic.
The plague is one of the greatest scourges of humankind. The disease is caused by an infection with the bacterium Yersinia pestis (Y. pestis). Three major plague pandemics have been documented, each resulting in a devastating death toll and great human suffering. The Second Pandemic started with the Black Death (1347-1353), which killed a large proportion of the European population in only a few years. It continued for another four centuries and repeatedly hit the continent with local or regional outbreaks. When considering this longstanding presence of the plague in Europe, the easing and final disappearance of the disease at the end of the 18th century seems all the more puzzling.
Scientists from Germany and Latvia have now conducted an ancient genomic study in which they investigated human remains from two cemeteries in Riga, Latvia, that were used as a burial ground for plague victims in the 17th century. DNA was extracted from the teeth of 16 skeletons and subjected to sequencing.
The researchers were also able to reconstruct a complete pest genome from individual number 488. Credit: Guntis Gerhards
The scientists were able to detect traces of the pathogen Y. pestis in the remains of four individuals. In two of them the DNA was so well preserved that entire genomes of the pathogen could be analyzed. It turned out that the genomes from Riga differed in one essential aspect from genomes from the time of the Black Death: they had a lower number of a specific gene, the Pla gene. Pla is a so-called virulence factor, which is crucial for the transmission of the bacterium.
Julian Susat, Ph.D. candidate at the IKMB and first author of the study published in the journal Scientific Reports, reanalyzed previously examined DNA sequences of other strains that had occured after the Black Death and found the same pattern—they were all depleted in pla and therefore less infectious for humans.
In search of pathogens: In order to avoid contamination with modern DNA, the samples are examined in a clean room. Credit: Ben Krause-Kyora
This raises the question why the bacteria reduced the number of their pla genes during the course of the second pandemic. Susat speculates: "One possibility is that the bacteria had to adapt to a new environment after the Black Death. The depletion of pla could have led to a fitness advantage when infecting rodents, the natural host of Y. pestis."
For the leader of the study Ben Krause-Kyora, the results provide a possible explanation for the course of the pandemic: "The spread of the pla-depleted, and thus for humans less virulent, strains could at least partially explain the decline and ultimately the disappearance of the plague from Europe."
The study highlights how the application of modern genome sequencing technologies can help to unmask adaptive processes of pathogens during a pandemic and thus explain the course of the disease.
More information: Julian Susat et al. Yersinia pestis strains from Latvia show depletion of the pla virulence gene at the end of the second plague pandemic, Scientific Reports (2020). DOI: 10.1038/s41598-020-71530-9
Journal information: Scientific Reports
Provided by Kiel University