Characterizing COVID-19 antibodies for potential treatments
New research led by Caltech has characterized a multitude of antibodies to the virus that causes COVID-19 and identified those that are most effective at neutralizing the virus. Antibodies are proteins produced by the body in response to infection. Ultimately, it is hoped that potent antibodies like the ones described in this study can be given as treatment or prevention for COVID-19.
The study was led by Christopher Barnes, senior postdoctoral fellow in Biology and Biological Engineering and Howard Hughes Medical Institute Hanna Gray Fellow. The research was conducted in the laboratory of Pamela Björkman, the David Baltimore Professor of Biology and Bioengineering, and utilized Caltech's cryo-electron microscopy and protein expression facilities. A paper describing the research appears in the journal Nature.
An individual can produce hundreds or thousands of different variants of antibodies against any pathogen or foreign substance, including viruses, leading to a wide diversity of antibodies in a single person and in the human population. Some antibodies are better at blocking a virus than others. Think of the numerous ways that a boxer can engage with an opponent: a punch to the face is more likely to knock out the opponent than a glancing punch to the leg. When an antibody effectively renders a virus unable to infect cells—knocking it out, so to speak—it is called "neutralizing."
"An ideal treatment would be a combination or 'cocktail' of different antibodies that attack the virus in different, but still effective, ways," says Barnes. "With a combination of antibodies, it's less likely that a virus can evolve to escape them."
Since the start of the COVID-19 pandemic, Barnes and Björkman have been studying the antibodies of individuals who have recovered from the disease, in search of those that are the most neutralizing. They utilize structural biology techniques to image the interactions between SARS-CoV-2 proteins and individual antibodies.
Each individual SARS-CoV-2 virus has large, spiky protein structures on its surface that, in cross-section, make it resemble a cartoon sun with rays ("corona," the Latin word for "crown," is also the scientific term for the wisps of plasma that spread outward from the sun). At the tip of each of these so-called spike proteins is a tripod of protein regions, each called a receptor-binding domain (RBD). An RBD can flip from a "down" position on the spike to an "up" position, which reveals a hidden site called the receptor-binding motif.
The target where SARS-CoV-2 latches onto a human cell is called the angiotensin-converting enzyme 2 (ACE2) receptor. Normally, this cell-surface receptor functions to regulate blood pressure, but SARS-CoV-2 has co-opted it as a means to gain entry into cells in the lungs and other organs. The receptor-binding motif acts as a grappling hook, grabbing onto the ACE2 receptor.
Once the virus has attached to a cell, it can fuse with the cell's membrane and invade the cell, turning the infected cell into a factory to make new viruses. An antibody that could block the receptor-binding motif or prevent fusion using a different mechanism would thus be very effective in preventing the virus from entering cells.
Barnes and his team aimed to discover how antibodies interact with the spike RBDs, in both their open (RBD "up") and closed (RBD "down") conformations. In a paper previously published in Cell, they collaborated with the laboratory of Michel Nussenzweig at The Rockefeller University to study individual antibodies (so-called monoclonal antibodies) collected from people who had recovered from COVID-19. Using the collection of monoclonal antibodies discovered at Rockefeller by their collaborators, the Caltech team used microscopy techniques that are able to image proteins with single-atom resolution to discover precisely where the various antibodies bound to SARS-CoV-2 spike proteins.
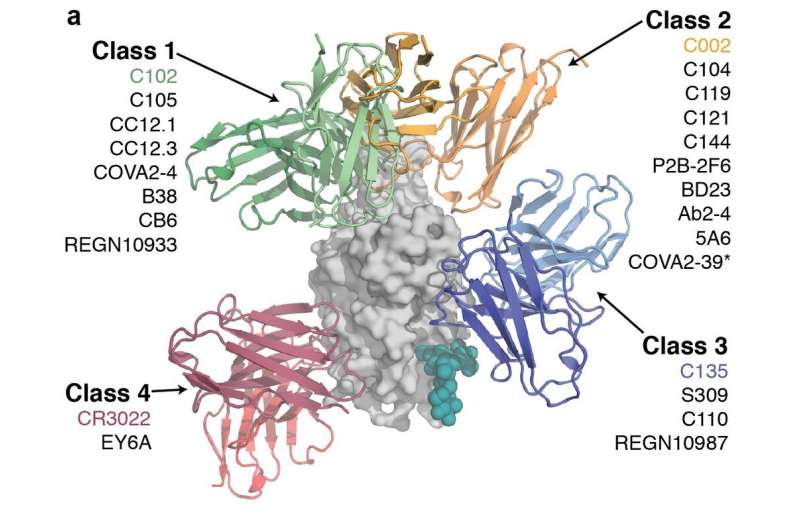
In their latest paper, Barnes worked with Björkman lab graduate students and microscopists from other Caltech labs to quickly solve eight new structures that show how neutralizing antibodies against SARS-CoV-2 block the RBDs on spike proteins to prevent the virus from entering cells. They found a variety of recognition modes: some antibodies bound spikes with three "up" RBDs, some bound to both "up" and "down" RBDs on the same spike, and some bound only to "down" RBDs.
From analyses of these structures, the team proposed four classes of anti-RBD antibodies based on whether they bound "up," "down," or both RBD conformations; whether their binding overlapped with the ACE2 binding site; and other criteria such as their potencies and derivation from particular antibody gene families. From these structures, the researchers proposed different mechanisms for virus neutralization.
For example, they found a particularly interesting antibody that binds simultaneously to adjacent RBDs, clamping all three RBDs into "down" positions that would lock the spike into a conformation that cannot open to expose the "grappling hook."
"First, I want to say how happy I am to see the high level of cooperation and collaboration within our lab and other Caltech labs to investigate these antibodies," Björkman says. "We think these structures will facilitate choices of the most effective combinations of monoclonal antibodies to be used for treatment of COVID-19 or to prevent viral infection of people in high-risk groups."
"In addition," she adds, "knowing the structures of these antibodies can facilitate the design of antibodies that bind more tightly to RBDs, thereby increasing their efficacy and lowering the dose needed for treatment. And finally, mapping where these antibodies bind is necessary information for structure-based design of vaccines to elicit the most potent classes of neutralizing antibodies."
"It's really great to have had the opportunity to work on a project that is so immediately relevant to a health crisis that affects the whole world," says Claudia Jette, a fifth-year graduate student and second author on the paper. "This is also the most collaborative project I've ever been a part of. Things moved fast and everyone went above and beyond to get things done, especially Christopher who was the lead on the project. It's been an absolute joy to work on something like this with the brilliant people that are in my lab."
"Our work provides the basis for future studies into patient-derived neutralizing antibodies from recovered COVID-19 individuals," says Barnes. "In collaboration with the team at Rockefeller, we are now working on characterizing temporal changes in antibodies isolated from the same donors. We hope that this future work will aid in our understanding of the potential for long-term protection against SARS-CoV-2 infection."
The paper is titled "SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies."
More information: Christopher O. Barnes et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies, Nature (2020). DOI: 10.1038/s41586-020-2852-1