The line of succession in neuron function
A specific region of messenger RNAs, the 3' untranslated region (3'UTR), plays an important role for cells to function properly. During embryonic development, 3'UTRs in hundreds of RNAs lengthen exclusively in neurons, which is crucial for the cells of the brain to function properly. The lab of Valérie Hilgers at the Max Planck Institute of Immunobiology and Epigenetics in Freiburg was now able to show that in Drosophila, an elegant and robust mechanism secures this process of 3'UTR extension. In their newest study they describe for the first time a hierarchical mechanism of take-over of function starting from the protein ELAV as a Master Regulator of neuronal transcript signatures, with a second-in-command protein FNE that is only activated if the Master is absent.
Neurons, also known as nerve cells, send and receive signals in our brain. They are particularly complex and unique in many aspects: Neurons do not divide or regenerate like many other cell types, and they can be excitatory, inhibitory or modulatory in their effect and motor, sensory or secretory in their function.
RNA molecules play an important role in making sure that neurons function properly. Nearly every known neurological or neurodegenerative disease in humans is associated with the malfunction of RNA regulation. During gene expression, messenger RNA molecules (mRNAs) are transcribed from the DNA and are later translated into a protein. But RNA is not simply the intermediate between DNA and protein. RNA has important regulatory roles.
Several regions of the mRNA molecule are not translated into a protein, for example the 3' untranslated region (3'UTR) that immediately follows the protein-coding region of the mRNA after the stop codon. Interestingly, the 3'UTR has a lot of regulatory roles on its own. For example, it can determine when and where a gene is active, dictates where in the cell a protein finds its place, and plays a particular role in synapse formation and maintenance of memory.
A unique RNA mechanism, which was discovered only a decade ago, distinguishes the nerve cells from all other cells: the addition of a long stretch of non-coding sequence to many mRNAs. This phenomenon, termed "3'UTR extension", is of crucial importance for neurons to maintain their unique identity, and to function properly.
The lab of Valérie Hilgers at the MPI of Immunobiology and Epigenetics in Freiburg focuses on this distinctive RNA processing mechanism. In previous work from the lab, it was found that in Drosophila, the protein ELAV is important for promoting 3'UTR extension in neurons. In this study, the team set out to learn more about ELAV.
ELAV as Master regulator of neuron biology
It turned out that ELAV is a so-called "Master regulator." In Biology, Master regulators are proteins that are deemed responsible for the control of a whole regulatory program of the cell. Thus, they are in charge of important, transformative events, on their own. Without them, the transformative event cannot take place. Relying on one single effector for such important tasks puts the cell at risk; this is why usually, several proteins share the work in biological systems.
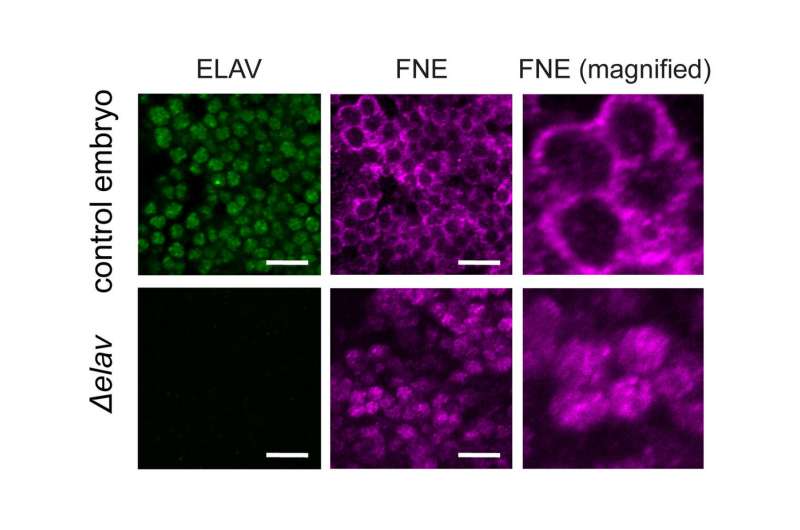
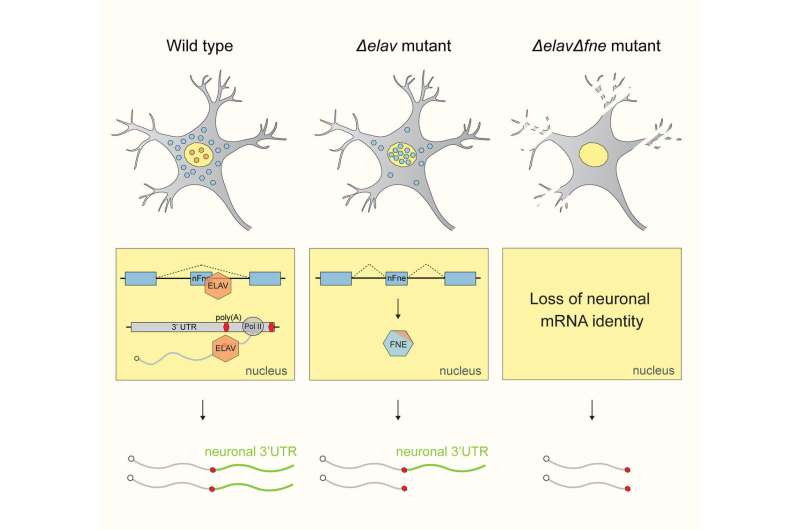
But the scientists found an unusual system of robustness that ensures the formation and maintenance of neuronal transcript signatures in Drosophila. "Under normal conditions, ELAV acts as a Master regulator and single-handedly mediates 3'UTR extension. Interestingly, it also represses the expression of the exon of another gene of the same family called FNE. But if we put the system under stress by mutating ELAV, we could show that a new, activated form of FNE is expressed, which translocates into the nucleus to perform the function of ELAV in its absence," says Judit Carrasco, first author of the study.EXAR mechanism manages succession
The team termed this hitherto unknown mechanism EXAR (EXon-Activated Rescue). "Considering the importance of "3'UTR extension" for the proper functioning of neurons, it makes sense to use such a robust mechanism, where succession is clearly defined," says Valérie Hilgers, who is also member of CIBSS—Centre for Integrative Biological Signalling Studies, Cluster of Excellence at the University of Freiburg.
This new insight into ELAV and the EXAR mechanism is a promising avenue for future research. ELAV is one of the best-conserved proteins in animals; it is found in every nerve cell of every animal. In humans, for instance, ELAV proteins are involved in numerous neurological diseases. The concrete mechanisms of how ELAV influences the health of neurons are not quite clear yet. The Hilgers lab in Freiburg wants to understand how ELAV proteins act on RNA to keep a neuron healthy.
More information: Judit Carrasco et al, ELAV and FNE Determine Neuronal Transcript Signatures through EXon-Activated Rescue, Molecular Cell (2020). DOI: 10.1016/j.molcel.2020.09.011