New tool enables easy, effective disease tracking
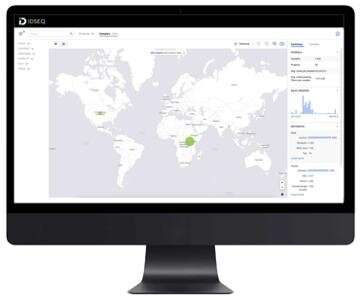
A new open source, cloud-based tool called IDseq can rapidly detect, identify and track emerging pathogens such as SARS-CoV-2. It can identify pathogens before there is an available complete genome sequence, and can therefore be used for current infectious disease outbreaks and emerging diseases. The developers have published a related article in the journal GigaScience
The coronavirus pandemic demonstrates the importance of global infectious disease monitoring. Finding the cause of an infectious disease outbreak is challenging, especially if it stems from a previously unknown pathogen. IDseq, an open source, cloud-based metagenomic analysis platform, identifies both novel and existing disease-causing pathogens from a human, animal or parasite sample to provide an actionable report of what is happening on the ground in labs and clinics anywhere in the world.
"IDseq can be thought of as an early warning radar for emerging or novel infectious agents," said Joe DeRisi, Ph.D., co-president of the Chan Zuckerberg Biohub. He contributed to the identification of the SARS coronavirus in 2003 and his research lab at the University of California, San Francisco, initiated the IDseq tool. It is designed to enable the global health community to leverage the ever-decreasing cost of sequencing for tracking and identifying infectious disease in essentially any sample.
"At the beginning of the coronavirus pandemic, researchers in Cambodia used IDseq to help confirm and sequence the whole genome of the country's first case of COVID-19 in a matter of days, and in California, we're providing critical SARS-CoV-2 genomic data to public health officials to inform contact tracing and intervention strategies," says DeRisi.
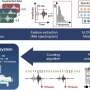
In their study, the scientists used various approaches to demonstrate that the IDseq tool can reliably identify emerging pathogens, among them, as proof of principle, a nasal swab from a COVID-19 patient in Cambodia. A partnership between the Chan Zuckerberg Biohub, the Chan Zuckerberg Initiative (CZI), and the Bill and Melinda Gates Foundation enabled the researchers to sequence and confirm the country's first case of COVID-19 in a matter of days—not the weeks it could typically take. The results demonstrate that IDseq can detect the presence of an emerging pathogen prior to the existence of a full reference genome. IDseq also now contains a new workflow for building SARS-CoV-2 consensus genomes.
"Metagenomic sequencing (mNGS) is an incredibly useful tool for pathogen detection because of its highly sensitive and hypothesis-free nature," said Katrina Kalantar, computational biologist at CZI. "We've seen labs that are using IDseq for existing mNGS studies rapidly pivot their focus to more targeted sequencing of SARS-CoV-2, which has helped researchers better understand coronavirus transmission patterns."
In Cambodia, researchers uploaded the genome sequence to the open source pathogen data repository Global Initiative on Sharing All Influenza Data and to Nextstrain. Using the system, scientists anywhere can see the full genome sequence of the SARS-CoV-2 coronavirus and study it within the broader context of SARS-CoV-2 coronavirus sequences uploaded globally. Researchers at the Cambodian National Center for Parasitology, Entomology and Malaria Control (CNM) and the National Institute of Allergy and Infectious Diseases (NIAID) partnered with the Institut Pasteur Cambodia to complete this research.
Unlike tests that are specific for a known agent, such as the SARS-CoV-2 PCR test, mNGS is a universal method that can detect novel disease-causing pathogens, which can be especially useful in cases where researchers may not know what is causing an infection, or what pathogens are circulating in a particular area. An mNGS experiment starts with mass-amplifying DNA traces of pathogens from a patient's sample, resulting in millions of small bits of DNA sequences, or reads. This enormous dataset must then be analyzed and interpreted using bioinformatic techniques. The aim is to assign individual DNA fragments from the clinical sample to specific pathogens by leveraging knowledge from sequence databases.
Analyzing the massive amount of data from a typical mNGS experiment often requires a battery of specialized bioinformatic tools, including highly specialized expertise and expensive commercially licensed software—making mNGS a hard-to-access method. The new user-friendly IDseq software is open source and freely available to the global health community, reducing the barrier of entry to metagenomics. Researchers can reuse and build upon the code, which works via a cloud-based service and a web application designed for collaboration and data sharing. The pipeline starts with raw sequencing data as the input, and then goes through steps of filtering, quality control, alignment, and reporting and visualization.
More information: Katrina L Kalantar et al. IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring, GigaScience (2020). DOI: 10.1093/gigascience/giaa111