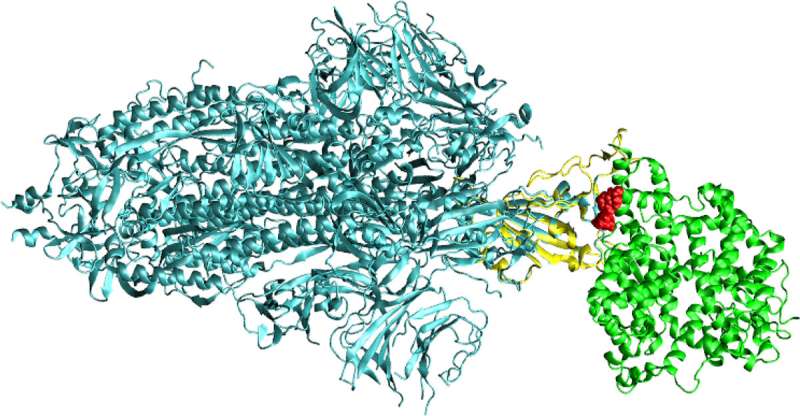
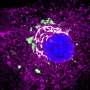
The structure of full spike protein (shown in cyan color) and ACE-2 (shown in green color) complex with a ligand (shown in red color) bound to the interfacial region of these two proteins. The receptor binding domain of spike protein is shown in yellow color. Credit: KTH Royal Institute of Technology
A virtual screening of the DrugBank database has identified a variety of as yet unexplored ways to attack SARS-CoV-2, even as it mutates. The study identified drugs and possible cocktails that are shown to target vital proteins of the coronavirus.
Recently publishing in the journal Scientific Reports, researchers from KTH Royal Institute of Technology, in collaboration with Alagappa University (India), proposed a list of individual drugs and cocktails that deserve testing for the treatment of COVID-19.
The researchers at KTH tested their own screening protocol, as an alternative to software typically used in the biopharmaceutical industry to screen vast volumes of compounds in drug databases.
Theirs involved a double-scoring approach to identify lead compounds that show potential for COVID-19 therapy. The procedure appears to have succeeded in avoiding false-positives—a common problem in virtual screening.
A key part of the study is the identification of drugs that target—or bind to—multiple proteins that are essential for replication of the virus, and which are also involved in the initial stage of host-cell infection. Corresponding authors Vaibhav Srivastava and Arul Murugan say multi-targeting offers an effective route to deal with drug resistance, which would enable a drug to work around mutations of the virus.
The researchers at KTH tested their own screening protocol which appears to have reduced false positive matches.
Mobile COVID-19 testing site in Sweden. Credit: David Callahan/KTH Royal Institute of Technology
"The virus is mutating rapidly, which means that it is modifying its proteins," Srivastava says. "If we have a drug that can target several proteins, and if one becomes mutated, the drug will be effective on others."
This attribute allowed the team to propose cocktails that have versatility. "It was possible for us to propose cocktails, or blends of drugs, in which each drug can bind to a specific target protein with high affinity," he says.
For example, the study proposed one cocktail, baloxavir marboxil, natamycin and RU85053, which targets the three viral proteins respectively, 3CL Main protease, papain-like protease and RdRp. Such drug cocktails have proven effective in the treatment of other virally-transmitted diseases, such as HIV.
Murugan says that the reliability of their approach was validated by the fact that the screening also identified drugs that are already in clinical trial. Furthermore, he says that such studies can provide valuable insights regarding why certain drugs were found to be ineffective. For example, they state that the drug hydroxychloroquine was non-effective mainly due to its poor binding affinity towards viral proteins.
Other drugs that the study recommended for testing were tivantinib, olaparib, zoliflodacin, golvatinib, sonidegib, regorafenib and PCO-371.
The paper also provides a listing of multi-targeting drugs such as DB04016, phthalocyanine, tadalafil, which can also be effective in combating the rapidly-mutating coronavirus.
More information: Murugan, N.A., Kumar, S., Jeyakanthan, J. et al. Searching for target-specific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach. Scientific Reports 10, 19125 (2020). doi.org/10.1038/s41598-020-75762-7
Journal information: Scientific Reports
Provided by KTH Royal Institute of Technology