Credit: CC0 Public Domain
AI-based evaluation of medical imaging data usually requires a specially developed algorithm for each task. Scientists from the German Cancer Research Center (DKFZ) have now presented a new method for configuring self-learning algorithms for a large number of different imaging datasets—without the need for specialist knowledge or very significant computing power.
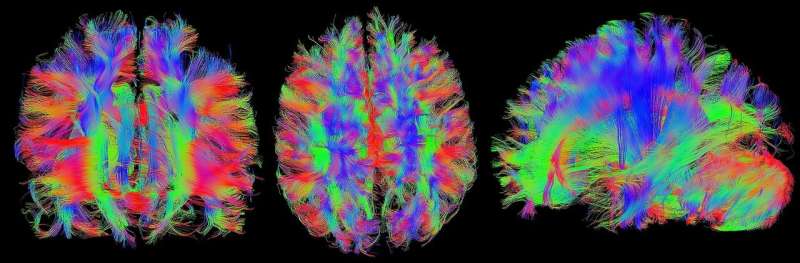
In the evaluation of medical imaging data, artificial intelligence (AI) promises to provide support to physicians and help relieve their workload, particularly in the field of oncology. Yet regardless of whether the size of a brain tumor needs to be measured in order to plan treatment or the regression of lung metastases needs to be documented during the course of radiotherapy, computers first have to learn how to interpret the three-dimensional imaging datasets from computed tomography (CT) or magnetic resonance imaging (MRI). They must be able to decide which pixels belong to the tumor and which do not. AI experts refer to the process of distinguishing between the two as semantic segmentation.
For each individual task—for example recognizing a renal carcinoma on CT images or breast cancer on MRI images—scientists need to develop special algorithms that can distinguish between tumor and non-tumor tissue and can make predictions. Imaging datasets for which physicians have already labeled tumors, healthy tissue, and other important anatomical structures by hand are used as training material for machine learning.
It takes experience and specialized knowledge to develop segmentation algorithms such as these. "It is not trivial and it normally involves time-consuming trial and error," explained medical informatics specialist Fabian Isensee, one of the lead authors of the current publication. He and his colleagues in the DKFZ division headed by Klaus Maier-Hein have now developed a method that adapts dynamically and completely automatically to any kind of imaging datasets, thus allowing even people with limited prior expertise to configure self-learning algorithms for specific tasks.
The method, known as nnU-Net, can deal with a broad range of imaging data: in addition to conventional imaging methods such as CT and MRI, it can also process images from electron and fluorescence microscopy.
Using nnU-Net, the DKFZ researchers obtained the best results in 33 out of 53 different segmentation tasks in international competitions, despite competing against highly specific algorithms developed by experts for specific individual questions.
Klaus Maier-Hein and his team are making nnU-Net available as an open source tool to be downloaded free of charge. "NnU-Net can be used immediately, can be trained using imaging datasets, and can perform special tasks—without requiring any special expertise in computer science or any particularly significant computing power," explained Klaus Maier-Hein.
So far, AI-based evaluation of medical imaging data has mainly been applied in research contexts and has not yet been broadly used in the routine clinical care of cancer patients. However, medical informatics specialists and physicians see considerable potential for its use, for example for highly repetitive tasks, such as those that often need to be performed as part of large-scale clinical studies. "NnU-Net can help harness this potential," study director Maier-Hein remarked.
The study is published in Nature Methods.
More information: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation, Nature Methods, DOI: 10.1038/s41592-020-01008-z , www.nature.com/articles/s41592-020-01008-z
Journal information: Nature Methods
Provided by German Cancer Research Center