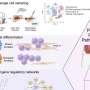
Graphical abstract. Molecular Cell (2020). DOI: 10.1016/j.molcel.2020.12.005
Transcription factors—proteins that regulate gene expression—play critical roles in cell fate decisions and are frequent targets of mutation in a variety of human cancers. Understanding how transcription factors contribute to disease requires the identification of their direct gene targets, but traditional methods are time-consuming and make it difficult to distinguish direct from secondary changes.
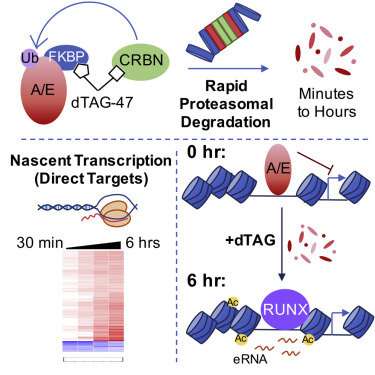
Kristy Stengel, Ph.D., Scott Hiebert, Ph.D., and colleagues have now devised a chemical genetic approach that speeds transcription factor analysis from days to minutes.
Reporting in Molecular Cell, the researchers applied the new approach to analysis of the fusion protein AML1-ETO, the most frequent chromosomal translocation in acute myeloid leukemia. They defined a small core network of about 60 direct AML1-ETO-regulated genes, including mediators of myeloid differentiation and cell fate decisions.
Their findings demonstrated that AML1-ETO repression of this small network impairs myeloid differentiation, and the mechanistic insights point to novel therapeutic strategies.
More information: Kristy R. Stengel et al. Definition of a small core transcriptional circuit regulated by AML1-ETO, Molecular Cell (2020). DOI: 10.1016/j.molcel.2020.12.005
Journal information: Molecular Cell
Provided by Vanderbilt University