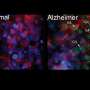
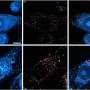
Researchers identify 'instigator' gene associated with Alzheimer's disease

In a new paper published in Nature Communications, Mayo Clinic researchers and collaborators report the protein-coding gene SERPINA5 may worsen tau protein tangles, which are characteristic of Alzheimer's disease, and advance disease. By combining clinical expertise, brain tissue samples, pathology expertise and artificial intelligence, the team clarified and validated the relevance of the gene to Alzheimer's disease.
The researchers used tissue samples from 385 brains donated to the Mayo Clinic Brain Bank, which houses more than 9,000 brain tissue specimens for the study of neurodegenerative disorders. The samples were from people who were diagnosed with Alzheimer's disease and lacked co-existing diseases found in the brain. This ensured a spotlight on Alzheimer's disease, which enabled the team to focus on targets relevant to the disease.
These samples were used to classify the pattern of protein tangles associated with Alzheimer's. Then the team used digital pathology and RNA sequencing to identify gene expression in the samples, which effectively measures gene changes responsible for instructing proteins.
"We were able to look at an entire disease spectrum and find gene changes that may really influence the hippocampus, the brain's memory center," says Melissa Murray, Ph.D., a Mayo Clinic translational neuropathologist and lead author on the paper. "That means we may have targets that indicate why some people have relative preservation and some people have relative exacerbation of memory loss symptoms."
Using a machine learning algorithm, the authors narrowed the genes of interest from about 50,000 to five. The top candidate, SERPINA5, was found to be strongly associated with tau tangle progression in the hippocampus and cortex of the samples. The researchers plan to investigate how SERPINA5 interacts with tau protein to develop an inhibitor.
"Much of the focus of therapeutics is on abnormal proteins—amyloid and tau—used to biologically define Alzheimer's disease," says Dr. Murray. "But we hope to take a step back to look at a new interacting partner that may be actually accelerating tau or pushing tau accumulation past the tipping point."
Based on a growing understanding of how Alzheimer's disease may affect people at different ages, or differences observed between women and men, the study team did not limit their investigation by adjusting for age and sex.
The team was recently awarded a grant by the state of Florida to ensure the findings are broadly applicable across an ethno-racially diverse Alzheimer's disease cohort, as the need for more research to clarify the relevance of gene expression changes on hippocampal vulnerability may be critical for preserving memory loss in Alzheimer's disease. But, Dr. Murray says, by starting with the human brain and ending with it, the researchers hope their findings provide a deeper level of understanding that helps advance findings to clinical trials faster.
"While we have direct evidence of SERPINA5 in the context of Alzheimer's disease, SERPINA3 in this same family of genes has also been looked at in Alzheimer's, and SERPINA1 in ALS. So I think it's about collective awareness and paying attention to this group of proteins."
More information: Angela M. Crist et al, Transcriptomic analysis to identify genes associated with selective hippocampal vulnerability in Alzheimer's disease, Nature Communications (2021). DOI: 10.1038/s41467-021-22399-3