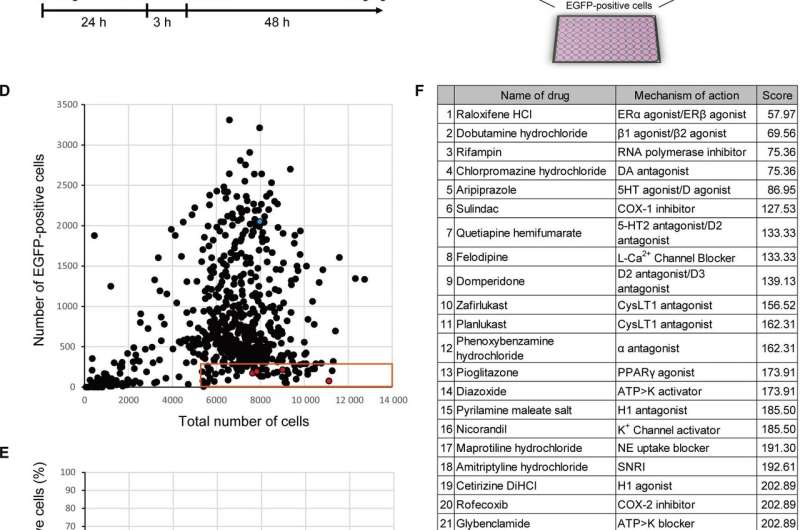
Compound screening using SeV and iPSCs. (A) Construction of SeV. SeV carrying EGFP gene in the 3’ region of viral genomic RNA was constructed after removal of F gene. (B) Timeline of compound screening. (C) Schema of compound screening. (D and E) Result of first screening. Approximately 500 compounds were evaluated. Blue dot shows negative control (DMSO). Red dots show the hit drugs raloxifene, rifampin, pranlukast, zileuton, and pioglitazone. (F) List of hit compounds. The scores reflect the number of EGFP‐positive cells. Credit: FEBS Open Bio (2021). DOI: 10.1002/2211-5463.13153
The last year has seen intensive research around the world on SARS-CoV-2, the virus responsible for the COVID-19 pandemic. Despite several vaccines already available, the rapid mutation of the virus is causing concern that the infection will continue to spread. A new study led by CiRA Professor Haruhisa Inoue shows that induced pluripotent stem (iPS) cells can assist in finding effective drugs for RNA viruses.
HIV, Ebola virus, and now the SARS-CoV-2 pandemic, in the last half century, RNA viruses have been responsible for many of society's greatest health calamities. One of the great challenges in managing these viruses is their rapid mutation rate. Indeed, it is possible that as SARS-Cov-2 continues to mutate, it may need, like influenza, new vaccines annually.
Inoue and his colleagues proposed a drug screening using different types of RNA viruses and cells as a possible solution.
"A common therapeutic approach for RNA viruses, like penicillin for bacterial infections, is needed not only to treat current RNA viruses but also newly emerging ones," said Inoue. "Drugs that have efficacy in different combinations of RNA viruses and cells may have therapeutic potential for future RNA virus infections. We screened for approved drugs that prevented Sendai virus infection in iPS cells and used them to test if they could prevent Ebola virus and SARS-CoV-2 infection in different types of cells."
Where drug discovery involves the screening of thousands of chemicals for which we know little about the effects on humans, drug repurposing screens drugs for which the safety is already known. This approach may find drug candidates more quickly than drug discovery.
Sendai virus is an RNA virus commonly used in iPS cell research and, unlike Ebola virus and SARS-CoV-2, does not require high-level biosafety laboratories for the experiments.
From the screening of 500 drugs, five showed promise for further investigation, including Raloxifene, a drug for breast cancer and osteoporosis, and Pioglitazone, a drug for diabetes mellitus.
While iPS cells are susceptible to Sendai virus infection, they are missing a key receptor that obstructs infection by SARS-CoV-2.
"Research has shown that SARS-CoV-2 infects cells by binding to ACE2 [angiotensin-converting enzyme 2]. Target cells of SARS-CoV-2 express a higher level of this receptor, but iPS cells express a low level," explained Inoue.
Therefore, to investigate these drugs in Ebola virus and SARS-CoV-2 infection, the study switched the iPS cells to other human and monkey cells. Consistent with its effects on Sendai virus, Raloxifene was found to have anti-viral activity against Ebola virus and SARS-CoV-2.
Raloxifene is a selective estrogen receptor modulator (SERM), a group of drugs commonly used to treat breast cancer and postmenopausal symptoms like menopause. The scientists therefore explored other SERMS, finding three more that had inhibitory effects on Ebola virus and SARS-CoV-2 infection.
Pioglitazone, on the other hand, had milder benefits for preventing SARS-CoV-2 infection, but none on Ebola virus infection. However, combining the two drugs had a slight synergistic effect on SARS-CoV-2 infection.
"These drugs, with different efficacies for RNA viruses and cells, may have therapeutic potential for RNA virus infections, including newly emerging and mutated viruses," said Inoue.
More information: Keiko Imamura et al. iPSC screening for drug repurposing identifies anti‐RNA virus agents modulating host cell susceptibility, FEBS Open Bio (2021). DOI: 10.1002/2211-5463.13153
Provided by Kyoto University