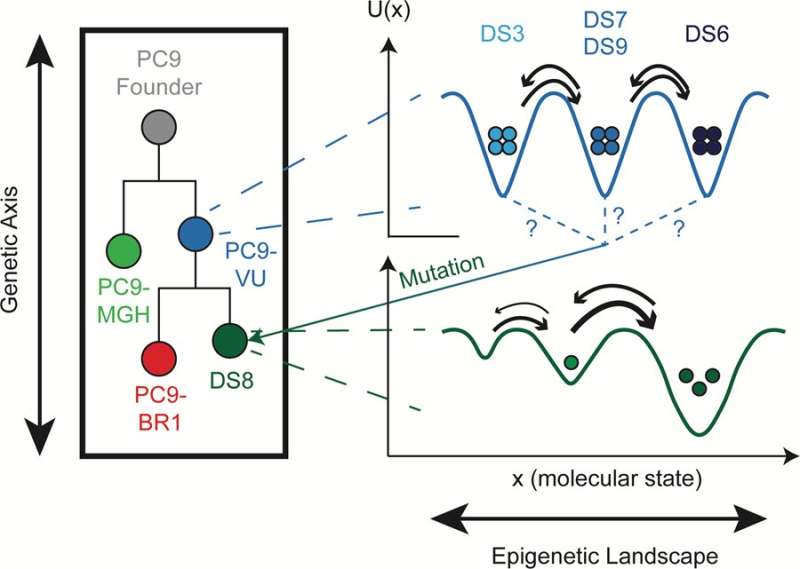
The figure above summarizes the findings of the team in terms of genetic, epigenetic and stochastic differences among the EGFR-mutant lung cancer cells studied. Credit: University of Arkansas
Leonard Harris, assistant professor of biomedical engineering, led a team of researchers from Vanderbilt University that has shown how an in vitro model of tumor heterogeneity, or diversity, resolves three different sources of cell state variability in cancer cells. The paper has been published in PLOS Biology.
A heterogeneous tumor is a tumor that is made up of many different types of cancer cells. Often, the cells have different types of genetic mutations and co-exist within a tumor. The diversity of the tumor is what makes cancer difficult to treat.
"It's like the success of a diverse team," Harris explains. "A team made up of people from different backgrounds, ages, stages of their career, etc., are often better at tackling problems because the team members provide different perspectives."
In a tumor, different cells respond to drug treatments differently. Some cells are able to survive and regrow the tumor and spread, which is why Harris and his team continue to research the ways surviving cancer cells differ from the other tumor cells.
But genetic mutations are not the only way cancer cells can differ from each other. Cells that have the exact same DNA can exist in very different states. For example, your skin cells and your liver cells have exactly the same DNA but they function very differently; that is an example of epigenetic heterogeneity. Moreover, when a skin cell divides, it produces two skin cells. The cells do not inherit the skin cell state from the DNA; it has to come through some other means. It is this non-genetic form of inheritance that makes the process epigenetic.
Cancer cells also differ due to random fluctuations in molecule numbers inside each cell: molecules randomly interact with each other, degrade, are synthesized by the cell, secrete into and out of the cell, etc. This type of non-genetic heterogeneity is called stochastic variability and is not heritable, unlike epigenetic processes. It might not seem like a big deal, but researchers have shown that stochastic variability can have major effects.
The experimental and computational work reported in the paper was performed at Vanderbilt University in collaboration with Corey E. Hayford, Darren R. Tyson, C. Jack Robbins III, Peter L. Frick and Vito Quaranta and has motivated many additional research projects. It is now the foundation for Harris' U of A laboratory.
"Cancer is commonly referred to as a 'genetic disease," meaning it is caused by mutations in critical parts of the DNA that cause cells to grow out of control," Harris said. "This has led to decades of research on the genetics of cancer, which has resulted in significant advances, including the development of numerous therapeutic drugs that target so-called 'driver oncogenes." While exceptionally effective in the short term, these targeted drugs fail almost universally, with patient tumors recurring within a few months to a few years. This has led many researchers to begin considering the role of non-genetic processes in the response of tumors to drugs."
Modeling and experimental techniques were used to distinguish the three different sources of variability among lung cancer cells: genetic, epigenetic and stochastic. As stated above, epigenetic and stochastic variabilities are different types of non-genetic variability. Epigenetically distinct cells look different, like the skin and liver cells from the example above, whereas stochastically distinct cells appear nearly identical but may act completely different.
"Distinguishing genetic from non-genetic, and epigenetic from stochastic, factors in drug response is crucial for developing new therapies that can kill tumor cells before they have a chance to acquire genetic resistance mutations," Harris said. "They all contribute to tumor drug response in different ways."
A framework for distinguishing genetic and non-genetic sources of heterogeneity in tumors has been proposed previously but is not yet widely accepted within the cancer research community because of a lack of strong experimental evidence. The team's paper provides strong support for this framework.
The analysis presented in the paper was applied specifically to EGFR-mutant non-small cell lung cancer. Harris' lab is currently applying these ideas to other cancer types as well, including small cell lung cancer, melanoma and bone-metastatic breast cancer.
"In my laboratory, we are working on building computational models of the molecular networks within cancer cells that give rise to the different epigenetic states, across which cells can transition to survive drug treatments," Harris said. "The long-term goal of my lab's research is to expand these models until they are of sufficient detail to act as virtual platforms for testing the effects of various drugs and identifying novel drug targets."
By constructing these so-called "digital twins," the hope is to one day use them to perform virtual drug screens on models built from samples of real patient tumors and then design personalized treatment options for those patients. This will require forming collaborations with bioinformaticians, experimentalists and clinicians here at U of A, the Winthrop P. Rockefeller Cancer Institute at the University of Arkansas for Medical Sciences in Little Rock, and elsewhere. "Hopefully, the publication of this paper will help spark some of those collaborations," Harris said.
More information: Corey E. Hayford et al, An in vitro model of tumor heterogeneity resolves genetic, epigenetic, and stochastic sources of cell state variability, PLOS Biology (2021). DOI: 10.1371/journal.pbio.3000797
Journal information: PLoS Biology
Provided by University of Arkansas