Credit: Unsplash/CC0 Public Domain
Immunology and bioinformatics researchers from The University of Queensland have identified a powerful tool for analysing large patient datasets. Their work could lead to better patient stratification, and the precise and quicker adoption of targeted therapies.
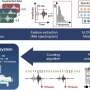
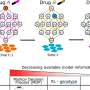
Led by UQ Diamantina Institute's Professor Di Yu and Dr. Yang Yang, who are based at The Translational Research Institute, the researchers compared four different mainstream tools to analyse patients' blood signatures, based on their gene expression. The methods were compared against 71 clinical datasets, each containing more than 100 patient samples.
"When you consider that we're looking at large datasets of patients, who each have more than 10,000 genes, we need a really good method to reduce the complexity of this big data for better interpterion," says Professor Yu.
"Of the four tools we compared, UMAP stood out as incredibly powerful. It performed significantly better than PCA, which is the tool many clinicians currently use to try and stratify patients," he says.
UMAP was the most efficient at reporting patient clustering. Using the tool, the researchers were able to separate healthy samples from those with lupus, as well as segregate the lupus patients into disease subgroups. They could also show which patients were getting better and who were getting worse.
The UMAP tool is relatively new, and is currently only used in biomedical research; however, Professor Yu hopes the publication of his team's results in the journal Cell Reports will lead to its future adoption in the clinic.
"UMAP's algorithm is based more on machine learning, and this makes it much more powerful than the popular PCA tool, which has a linear approach," says Professor Yu.
More information: Yang Yang et al, Dimensionality reduction by UMAP reinforces sample heterogeneity analysis in bulk transcriptomic data, Cell Reports (2021). DOI: 10.1016/j.celrep.2021.109442
Journal information: Cell Reports
Provided by Translational Research Institute