Relieving pain by mapping its biological signatures
Many people are confronted with chronic pain that can last for months or even years. How to best treat chronic pain? First, pain must be categorized for the right treatment to be prescribed. However, is that it is very challenging for patients to define their pain, its intensity or even its location using questionnaires. To overcome this difficulty, scientists from the University of Geneva (UNIGE) have joined forces with the research department of the Clinique romande de réadaptation (CRR) in Sion to carry out a complete epigenomic analysis of patients, making it possible to find the epigenetic signatures specific to each pain category. Thus, a simple blood test would make it possible to define which pain the person suffers from and, in the future, to prescribe treatment accordingly and to observe whether the biomarkers modified by the pain return to normal. These results can be read in the Journal of Pain.
Chronic pain is classified into two main categories: nociceptive pain—defined by the activation of receptors at the end of nerve fibers and found in osteoarthritis, burns or infections—and neuropathic pain, which is caused by damage to nerve structures, such as pain caused by shingles. In order to classify which pain the patient suffers from, they fill in several questionnaires and quantify pain intensity of using assessment scales. However, this is very subjective and time-consuming.
Blind genome analysis
"At the CRR, we treat many people suffering from chronic diseases," explains Bertrand Léger, a researcher at the CRR and last author of the study. "We joined forces with UNIGE scientists to carry out a complete epigenomic study and define specific biomarkers for each type of pain, in order to be able to categorize the various types of pain quickly and reliably."
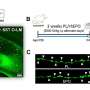
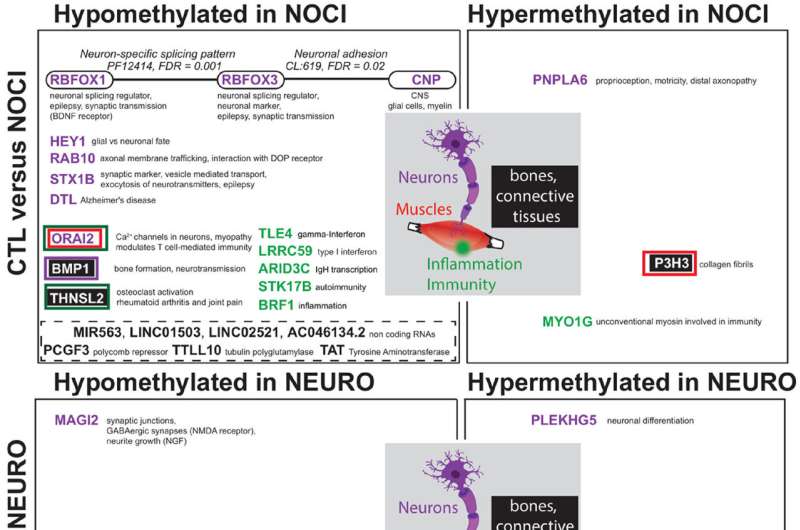
To do this, the Geneva team carried out an analysis of the entire genomes of 57 patients: 20 with no pain, 18 with nociceptive pain and 19 with neuropathic pain. "The aim was to start without any prior hypothesis to probe the genome as a whole and identify all the biomarkers involved in pain," explains Ariane Giacobino, the study's coauthor and a professor in the Department of Genetic Medicine and Development at UNIGE Faculty of Medicine.
Specific and potentially reversible biomarkers
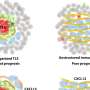
Unexpectedly, not only did the scientists identify very striking epigenetic signatures of pain, but there was no overlap between nociceptive and neuropathic pain. "This total absence of similarities between the two categories of pain is very surprising, because intuitively, we might think that the difficulty in defining one's pain comes from a similarity in the epigenetic signature. We could prove that itis absolutely not the case," notes Ariane Giacobino.
Indeed, the biomarkers specific to nociceptive pain are expressed by the genes of the opioid system—involved in emotion, reward and pain—as well as by the genes of inflammation, specific to irritation. Conversely, the biomarkers for neuropathic pain are linked only to genes of the GABA system, the neurotransmitters of the central nervous system.
"Now that these epigenetic signatures are clearly defined, a simple blood test will make it possible to define the type of pain the person is suffering from and prescribe the appropriate treatment," says Bertrand Léger. The treatment will thus no longer target the symptoms, but the very root of the problem. And finally, since epigenetics is characterized by the fact that the expression of a gene is durably modified, the right treatment may return it to normal. "We could imagine monitoring the reversion of pain by observing, from an epigenetic point of view, whether the biomarkers return to normal, and adapt the treatment accordingly," concludes Ariane Giacobino.
More information: Ludwig Stenz et al, Genome-wide epigenomic analyses in patients with nociceptive and neuropathic chronic pain subtypes reveals alterations in methylation of genes involved in the neuro-musculoskeletal system, The Journal of Pain (2021). DOI: 10.1016/j.jpain.2021.09.001