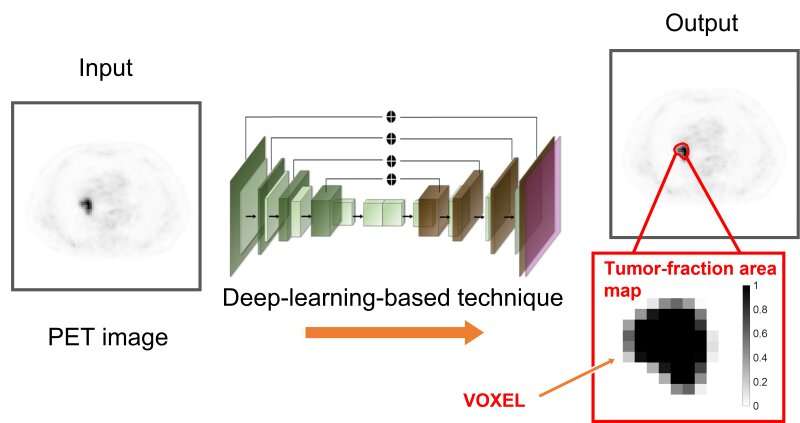
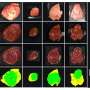
In the tumor-fraction area map each square is a voxel. The deep learning technique can determine how much of a gray area in each voxel is tumor or normal tissue (see scale on right from 0, no tumor to 1, all tumor). The technique is designed to accurately calculate the volume of tumors on PET images with the aim of improving patient care. Credit: Abhinav Jha, Washington State University in St. Louis
NIBIB-funded engineers are using deep learning to differentiate tumors more accurately from normal tissue in positron emission tomography (PET) images. Standard analysis of PET scans count black regions as tumor and white regions as normal. The team at Washington University in St. Louis has developed a technique using statistical analysis and deep learning to determine the extent of tumors at their margins where the images show various shades of gray.
PET images consist of what are known as voxels, which are 3-dimensional pixels in space. Current methods count these gray regions as either tumor or normal.
"The key idea is that we don't just learn if a voxel belongs to the tumor or not," said team leader Abhinav Jha, Ph.D., assistant professor of biomedical engineering in the McKelvey School of Engineering Jha said. "The voxel can be part tumor and part normal. The novelty is that we can estimate how much of the voxel is the tumor."
The research aims to provide more accurate information about the tumor to guide treatment decisions and improve patient care.
"It's a quality-of-life issue for patients," said Jha. "Helping to answer those questions would be satisfying and rewarding."
The work is reported in the journal Physics in Medicine & Biology.
More information: Ziping Liu et al, A Bayesian approach to tissue-fraction estimation for oncological PET segmentation, Physics in Medicine & Biology (2021). DOI: 10.1088/1361-6560/ac01f4
Journal information: Physics in Medicine and Biology
Provided by National Institutes of Health