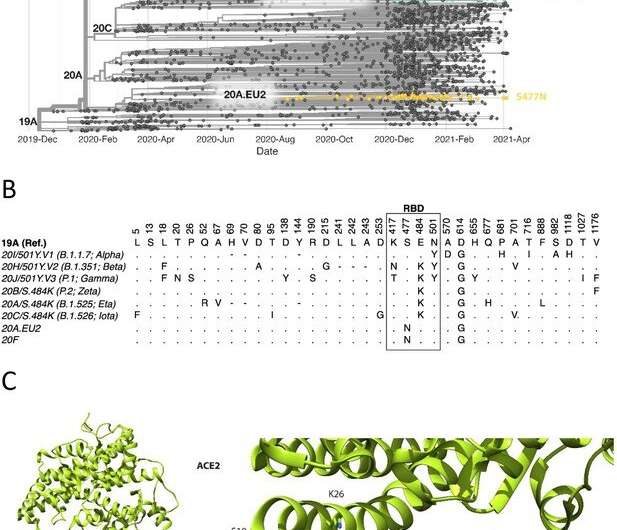
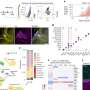
Spike RBD and ACE2 variants analyzed in this study. (A) Phylogenetic tree illustrating the clades containing the RBD mutations investigated in this study. Constructed using TreeTime (Sagulenko et al., 2018) from the Nextstrain Global (Hadfield et al., 2018) sample of SARS-CoV-2 sequences from the GISAID database (Shu and McCauley, 2017) (accessed 15 April 2021, N = 4017). (B) Alignment illustrating the Spike residues that differ between SARS-CoV-2 variants, with the RBD mutants boxed. The variants are labeled with their clade designation from Nextstrain (Hadfield et al., 2018) and/or PANGO lineage (Rambaut et al., 2020), where relevant. The RBD mutations were collated from CoVariants (Hodcroft, 2021) and Nextstrain. (C) The structure of human ACE2 (green) in complex with SARS-CoV-2 Spike RBD (cyan). The area enclosed by the box is shown enlarged on the right, with the residues mutated in this study labeled. Drawn using UCSF Chimera (Pettersen et al., 2004) using coordinates from PDB 6m0j (Lan et al., 2020). Credit: DOI: 10.7554/eLife.70658
Researchers from the Universities of Oxford and Dundee have made a discovery that helps explain why variations in the virus causes COVID-19 to spread so rapidly.
Coronaviruses are so named because of the spikes on their surface that make it look like a crown, the Latin word for which is corona. The virus uses these spikes to attach to and enter cells, where they then replicate. All common SARS-CoV-2 variants have mutations in the part of their spike proteins that binds to cells.
The Oxford-Dundee team found that most, but not all, of the common mutations in spike individually strengthened binding to ACE2, a protein found on the surface of our cells.
Furthermore, ACE2 variants found naturally in humans were shown to strengthen binding between it and the virus, suggesting that individuals with common ACE2 variants could be more susceptible to COVID-19 infection.
Professor Anton van der Merwe, from the Sir William Dunn School of Pathology at the University of Oxford, said: "The purpose of our study was to measure the precise effect of mutations in spike and ACE2 on the strength of their interaction.
"This is important because it helps us understand why some SARS-CoV-2 variants spread more rapidly and should also help us predict whether individuals with mutations in ACE2 would be more susceptible to COVID-19.
"Knowing the precise effect of spike mutations on binding to ACE2 helps us understand why SARS-CoV-2 variants spread more rapidly. This may inform our response to these new variants and help us identify potentially dangerous new variants before they spread widely."
Even as the world recovers from the pandemic, more infectious variants of the coronavirus have emerged. The Alpha variant rapidly replaced all other variants in the UK and in other countries, while the Delta variant, which largely superseded it, continues to spread around the world. The Beta and Gamma variants also emerged in populations that had previously been infected with the original SARS-CoV-2 virus.
This has led scientists around the world to study these new variants to try and understand the properties which make them more infectious. This latest research has also shown that while common ACE2 mutations led to increased binding, this is not the case for all SARS-CoV-2 variants.
Professor Geoff Barton from Dundee's School of Life Sciences, said: "These results are a great example of how collaboration between experts in Computational Biology working with leading experimentalists can result in exciting new findings.
"The work sprang from a computational analysis carried out in our group at Dundee by Dr. Stuart MacGowan on the effects of human ACE2 variants on SARS-CoV-2 binding. This helped the Oxford group to focus their complementary skills in the laboratory on the most important human and coronavirus variants."
The paper is published today in eLife.
More information: Michael I Barton et al, Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics, eLife (2021). DOI: 10.7554/eLife.70658
Journal information: eLife
Provided by University of Oxford