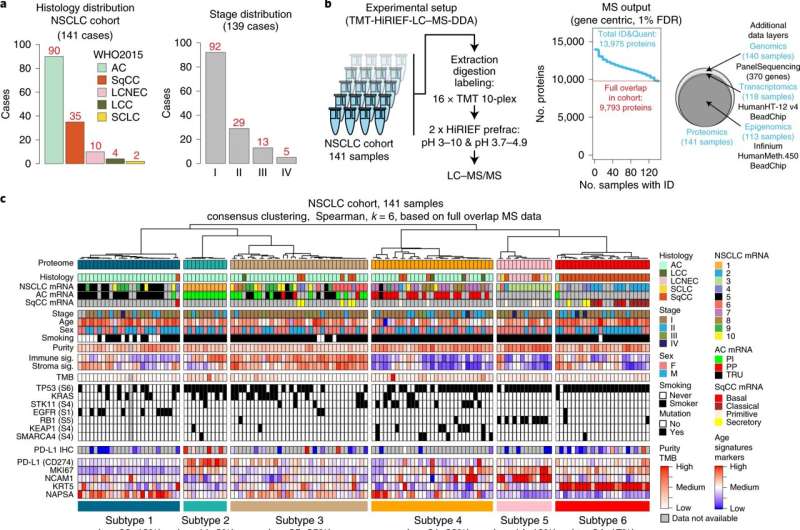
Fig. 1. MS-based identification of NSCLC proteome subtypes. a, Bar-plots showing histology and stage distribution in the patient cohort. b, Overview of experimental setup for MS-based proteome profiling, analysis output and supporting data levels. c, Hierarchical tree showing the results from consensus clustering used to identify NSCLC proteome subtypes. Annotation bars below indicate clinical information of samples, mRNA subtypes, infiltration signatures, common mutations and protein levels of selected markers. d, NSCLC proteome subtype network analysis with UMAP plot colored by modules (left), modules versus subtypes heat map (center) and cell types/signaling pathway enrichment analysis output for the ten modules (right). e, Box-plot indicating the number of overexpressed oncogenes per sample by NSCLC proteome subtype (n = 141 samples). Middle line, median; box edges, 25th and 75th percentiles; whiskers, most extreme points that do not exceed ±1.5× interquartile range (IQR). P value was calculated using Kruskal–Wallis test and number of samples per subtype is indicated in red. f, Bubble-plot indicating CDRPs commonly overexpressed in NSCLC cohort. OG, oncogene; TSG, tumor suppressor gene; quant, quantifications. g, Scatter-plot indicating mRNA to protein Pearson’s correlation (corr) of CDRPs. The corresponding correlation density plot is displayed on top. h, Scatter-plot showing promoter methylation to mRNA correlation versus mRNA to protein correlation for CDRPs. Indicated (top and right) are the corresponding density plots for the full gene-wise overlap (9,018 genes). Dunn’s multiple comparison tests with Benjamini–Hochberg adjustment for box-plot (e) are available in Supplementary Table 3. Credit: DOI: 10.1038/s43018-021-00259-9
Analyzing all the proteins that exist in a tissue type (the so-called proteome) can provide vital information on the causes of diseases and how they can best be treated. We talk to Janne Lehtiö, professor at the Department of Oncology-Pathology, about proteome-based medicine and what it can contribute to personalized cancer therapy.
What is proteome-based medicine?
The proteome is a snapshot of all the proteins that exist in a cell or a particular tissue. Proteome-based medicine is about measuring them and using the information to learn about a disease and, for instance, guide the choice of treatment, says Janne Lehtiö, professor at the Department of Oncology-Pathology at Karolinska Institutet.
How is it different to genome-based medicine?
Genome-based medicine is about using information about the genome—DNA in a cell—for the purposes of diagnosis or choosing a therapy. But it is proteins that perform the functions in the cells, and proteins that are the targets for almost all drugs. This is why we need information about the proteins to understand the mechanisms behind health and disease.
What can you learn from the proteome data?
Many complex diseases, such as diabetes and cancer, are caused not by one but by multiple changes in the genome. Because of this, it's hard to figure out how they collectively cause the disease. But at a protein level, the effect of the multiple genomic changes is more apparent, which gives us a chance to understand how the disease can best be treated.
I work with proteogenomics, which involves combining information from both the genome and the proteome. For example, it can answer questions about how a mutation affects all the cell's proteins or activity of multiple drug's target proteins.
How far has research on proteome-based medicine come?
Techniques for simultaneously measuring a large number of proteins have been developed in the past five years, so that we can now measure the proteome in clinical samples. We know that the proteome provides interesting information and we're starting to see some exciting papers showing just this for different forms of cancer, amongst other diseases. To date, the research has mainly focused on analyzing already collected tumor material. We now have to start analyzing patient samples at the point of therapy decision in order to analyze how the information can impact clinical decision making in relation to the other data used for it.
How might this affect the care of cancer patients?
At the end of the day it can mean being able to choose the best therapy for the patient and avoiding treatments that you suspect to be ineffective, using what we know about this patient's proteome. I also believe that proteome-based medicine and proteogenomics will be crucial if we're to learn how best to combine cancer drugs into one effective cocktail.
You and your colleagues have published a paper in Nature Cancer in which you have studied the proteome and genome in lung tumor samples. What do your results show?
We've found that we can use the proteome to divide the tumors into new subgroups, which differ from each other in an interesting way. The tumors use a range of mechanisms to hide themselves from the immune system, and these mechanisms differ from group to group. This is valuable information since it determines whether the patient will be helped or not by certain immunotherapies, which are important modern cancer drugs.
What's next for your research?
We'll start to do proteome analyses for lung cancer patients admitted to Karolinska University Hospital. Our work will form part of the lung cancer pilot that will be started under Karolinska Institutet's taskforce for precision medicine.
To start with, we'll gather proteome data in parallel with other data types. We'll see how the patient responds to the treatments she receives and if we could have predicted this response more accurately from our research results on the proteome. This might be followed by a study in which the patient's proteome data gets to decide which treatments are given in order to see of this gives better response outcomes than the current means of choosing a therapy.
More information: Janne Lehtiö et al, Proteogenomics of non-small cell lung cancer reveals molecular subtypes associated with specific therapeutic targets and immune-evasion mechanisms, Nature Cancer (2021). DOI: 10.1038/s43018-021-00259-9
Journal information: Nature Cancer
Provided by Karolinska Institutet