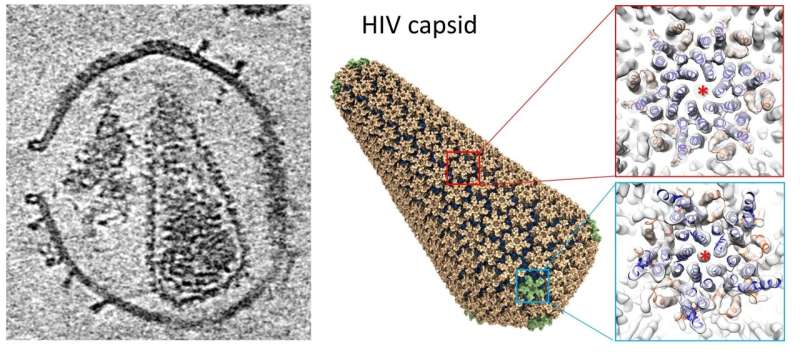
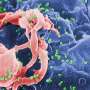
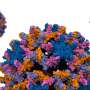
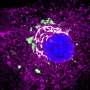
Structure of a HIV capsid. (Left) Central slice view of a HIV virus-like particle with pore-forming toxin on the membrane. (Middle) Atomic model of a HIV capsid. (Right) Density map of HIV capsid components. Credit: Science Advances
A new technique using electron tomography and subtomogram averaging at Diamond's electron Bio-Imaging Centre (eBIC), has solved the structure of the HIV capsid alone and in complex with host factors. This work also led to the building of an atomistic model of the whole HIV capsid using information gained from electron tomography, which the team believes could serve as a blueprint for the development of capsid-targeting antivirals.
The research paper, titled "High-resolution cryoET structures of native HIV-1 capsid in complex with IP6 and CypA" is published today (19 November) in Science Advances. The work was a collaboration between scientists at the University of Oxford, eBIC—the UK's national cryo-electron microscopy facility within Diamond Light Source and the University of Delaware.
The team was led by Professor Peijun Zhang, director of the eBIC at Diamond and Professor of Structural Biology at the University of Oxford. Lead author Dr. Tao Ni, University of Oxford, says, "Despite the global efforts to combat HIV/AIDS and the achievement of antiviral treatments, there are still approximately 38 million people with HIV/AIDS with no complete cure so far."
He explains that the human immunodeficiency virus (HIV) is a retrovirus; its RNA genome is encapsulated inside a conical-shaped capsid. During infection, HIV assembles and buds as immature virions with Gag polyprotein, which undergoes a maturation process, a step involving proteolysis and conformational change, which converts from an immature spherical shape to mature conical capsid. The capsid plays multiple essential roles during the early stage of HIV-1 replication, including protecting the genome from cellular innate immune responses and fostering reverse transcription, as well as regulating intracellular transport and entry into the nucleus. Many of these functions are affected by its interactions with host cell factors and small molecules.
However, because of the metastable property of HIV-1 capsid, isolating fully intact native capsids in quantities and concentrations suitable for high-resolution structural analyses has been challenging; the capsid suffers artefactual dissociation after the membrane is dissolved by detergent, a traditional method of capsid purification.
"To solve this problem, Peijun Zhang's team devised a novel approach. Instead of detergent extraction, we punctuate the membrane of HIV virus-like particles with a pore-forming toxin, which avoids the trauma associated with lysis of the virions and isolation of the cores, but also makes the capsid accessible to external cell factors and small molecules," says Dr. Ni.
Having established the experimental approach, the authors investigated the interactions between the authentic HIV capsid and a cellular factor Cyclophilin A (CypA), and a small-molecule cofactor IP6 (inositol hexakisphosphate). The team then applied electron tomography and subtomogram averaging to these samples.
Using this new technique, the team solved the structures of HIV capsid alone, and its complex with CypA and IP6 at around 5.4 Å resolution. These structures confirm the double IP6 binding site in the mature HIV capsid and provide insights into the role of IP6 and CypA in regulating HIV capsid stability.
Prof Zhang says, "In collaboration with Prof. Juan Perilla's group in the University of Delaware, using information derived from electron tomography, we also built an atomistic model of the whole HIV capsid which could serve as a blueprint for the development of capsid-targeting antivirals. The perforation of the enveloped virus membrane also provides a novel approach to study host-virus interaction for other viral systems."
More information: Tao Ni et al, Structure of native HIV-1 cores and their interactions with IP6 and CypA, Science Advances (2021). DOI: 10.1126/sciadv.abj5715. www.science.org/doi/10.1126/sciadv.abj5715
Journal information: Science Advances
Provided by Diamond Light Source