Credit: Pixabay/CC0 Public Domain
While considerable advances have been achieved in our battle against the coronavirus, new mutations of COVID19 continue to emerge and could threaten public health.
To prevent further severe pandemics, researchers from the University of Copenhagen and the immunotherapy company Evaxion have teamed up to develop a new AI tool that can more quickly and effectively predict how different protein elements can be assembled to increase the likelihood of coronavirus protection.
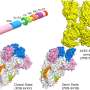
Their tool, BIFROST, is a computer model that uses algorithms to put together virus proteins that are most likely to be included in a vaccine, explains Christian Thygesen, an industrial Ph.D.
Thygesen has developed the model together with Evaxion and Associate Professor Thomas Hamelryck in the Deep Probabilistic Programming group at the University of Copenhagen's Department of Computer Science.
"For a vaccine to be effective, the body must be able to produce antibodies against viruses. It does so if it recognizes dangerous proteins—such as coronavirus spike proteins. With BIFROST, we use algorithms to prioritize the parts of viral proteins that we already know can stimulate an immune response, so that we can assemble them in a way that is most likely to work in a vaccine," says Christian Thygesen.
BIFROST uses data on amino acid chains—the building blocks of proteins—to predict how various proteins look and behave. In the future, this knowledge will allow researchers to design "super proteins" that elicit the desired response to viruses in the immune system, with few side effects.
BIFROST—faster, cheaper, better
BIFROST has numerous advantages over other models, according to a new study conducted by the three researchers.
Until now, researchers have used a computer model called Rosetta to learn about the shape and behavior of proteins. But as Christian Thygesen explains, the Rosetta method has significant shortcomings:
"Our new method has the major advantage of running on special hardware that allows us to get answers in seconds rather than waiting hours for results. It saves time and thus money."
BIFROST has another unique attribute that makes it more efficient than Rosetta.
"Where, based on a single amino acid chain, Rosetta can only provide one estimate of the protein in question, our tool uses algorithms to calculate the probability of several possible proteins. One piece of an amino acid chain doesn't have to result in exactly the same proteins every time," explains Thygesen
Thus, BIFROST is equipped to provide us with more suggestions about potential protein shapes and behaviors. This is important when trying to develop a vaccine that needs to be able to recognize many new variants of, for example, coronavirus spike proteins.
However, Evaxion's Anders B. Sørensen explains that there is still quite a way to go before the design behind BIFROST can be deoployed in a real vaccine.
"We have proven that BIFROST acts as a concept in the design phase, whereas real-world testing with animal models will be completed in 2022. So there remains a ways to go for us to test how our designed proteins function in humans. Nevertheless, with BIFROST, we have taken an important step towards creating a vaccine that can protect us from future pandemics," Sørensen concludes.
More information: Christian B. Thygesen et al, Efficient Generative Modelling of Protein Structure Fragments using a Deep Markov Model, bioRXiv (2021). DOI: 10.1101/2021.06.22.449406
Provided by University of Copenhagen