Could gene networks resembling air traffic explain arteriosclerosis?
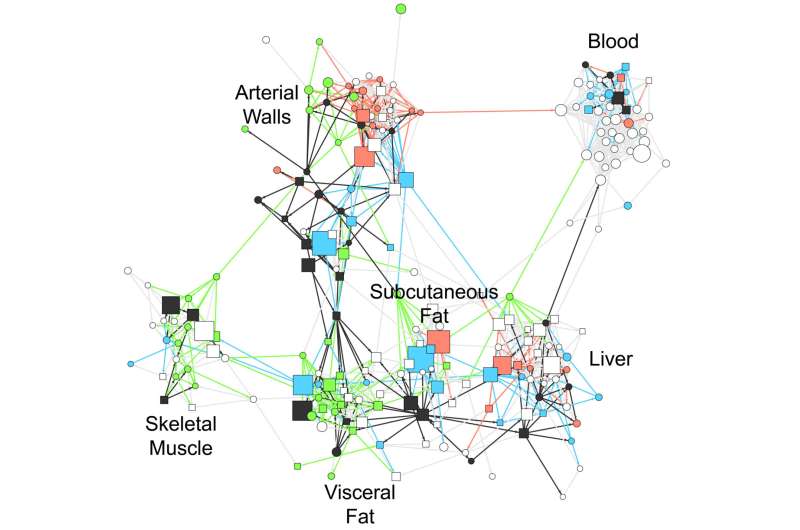
Up to 60 percent of the risk associated with coronary arteriosclerosis may be explained by changes in the activity of hundreds of genes working together in networks across several organs in the body. Moreover, fat processing hormones may play a central role in coordinating this activity. That is the primary result of a study that began nearly 20 years ago on a hunch and involved hundreds of coronary artery disease patients from Northern Europe. The study was led by researchers at the Icahn School of Medicine at Mount Sinai.
"It is well known that coronary artery diseases are driven by metabolic disorders. Our results suggest that much of this relationship is best explained by a complex series of multi-organ gene regulatory networks that are reminiscent of the hub and spoke maps used to depict global airline traffic," said Johan L.M. Bjorkegren, MD, Ph.D., Professor of Genetics and Genomic Sciences, and Medicine (Cardiology), and a senior author of the study published in Nature Cardiovascular Research. "We hope these network maps will give researchers the mechanistic framework needed to combat cardiovascular disease and develop more precise and personalized therapies."
Coronary artery disease results from a set of metabolic disorders which cause cholesterol and other factors to build up and clog a person's coronary arteries. This may lead to heart attacks or strokes. Affecting about 18.2 million Americans, coronary artery disease is the most common cause of heart disease in the United States. Risk factors, such as high cholesterol, high blood pressure, high blood sugar, and obesity, can involve a variety of organs. Although recent studies have shown that about 20 percent of the risk associated with this disease may be linked to slight differences in a person's DNA sequences, very little is known about how these differences may alter gene activity to produce coronary artery disease.
To address this issue, researchers studied the gene activity across seven different tissues found in the body. Tissue samples were obtained from 850 Estonian patients during open breast surgery. The patients were part of the Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task (STARNET) study. Six hundred of the patients had coronary artery disease whereas the other 250 did not. Tissue samples were collected by researchers in the lab of Arno Ruusalepp, MD, Ph.D., who is chief vascular surgeon at the Tartu University Hospital in Estonia.
Gene activity was analyzed from the following tissue samples: blood, liver, skeletal muscle, visceral abdominal and subcutaneous fat, and two pieces of the arterial wall taken from different parts of the heart. Dr. Bjorkegren began the study more than 20 years ago when he was training as a heart surgeon.
"Back then I had a hunch. Advances in genome sequencing and the Human Genome Project offered researchers the promise to fully understand the biology behind complex disorders. Scientists were showing how these disorders can be linked to dozens of tiny DNA sequence differences, most of which are not part of any gene codes. Thus, we needed a way to understand how these tiny but numerous DNA differences may actually cause metabolic disorders and coronary artery disease. These patients provided a unique opportunity to bridge this knowledge gap by allowing my team to measure gene activity in disease-relevant organs throughout the body," said Dr. Bjorkegren, who is also an associate professor at the Karolinska Institutet in Sweden.
Gene activity was determined by measuring the levels of RNA molecules in each tissue sample. These RNA molecules essentially contain photocopies of the DNA instructions for making life-sustaining proteins and other types of RNA molecules that are encoded in our genes.
Dr. Bjorkegren's team worked with researchers from around the world to test out the different ways that gene activity may be associated with the development of coronary artery disease. In particular, researchers in the lab of Jason Kovacic, MD, Ph.D., Executive Director at the Victor Chang Cardiac Research Institute in Darlinghurst, Australia, played an instrumental role.
Initial results supported previous findings that the activity of individual genes from certain tissues may be associated with a variety of cardiometabolic disorders as well as coronary artery disease. For instance, liver cells from coronary artery disease patients had greater changes in the activity of genes that control cholesterol production than those from control patients. However, these results did not fully explain how the activity of these genes worked together to cause coronary artery disease.
In contrast, most of the risk could be explained by different networks of disease-associated gene activity. Here the researchers used advanced computer programs to test out how the activity of all of the disease-related genes were grouped together in different combinations. Then, the validity of these networks were tested out using data from previously published reports.
In addition to the 20 percent identified by earlier studies, the results of the current study showed that an additional 54-60 percent of the risk associated with coronary artery disease could be explained by 224 of these gene regulatory networks and that many of these networks could help explain the status of arteriosclerosis severity in individual cases. Of those networks, 135 were located within one type of tissue whereas the remaining 89 represented coordinated gene activity across multiple tissues.
The multi-tissue networks appeared to have the greatest impact. On average they could explain three times more of the disease risk than the single-tissue ones. One example of a multi-tissue network, called GRN165, accounted for 4.1 percent of risk for coronary artery disease and involved 709 genes active in the arterial wall as well as the subcutaneous fat tissue.
"We found that gene networks work like airplane traffic patterns. Just as a delay at one airport in a key state can disrupt flights in the entire nation, we found that a slight change in the activity of key genes in one tissue can disrupt the activity of other genes throughout rest body," said Dr. Bjorkegren.
Finally, the analysis suggested that hormones which help fat cells communicate with other organs—particularly the liver—play a critical role in coordinating the multi-organ networks. Support for this idea was, in part, based on experiments in mice when injections of some of these hormones altered blood fat and sugar levels.
Dr. Bjorkegren's team has set up a website where researchers can test whether a candidate gene may be part of these networks (starnet.mssm.edu).
"Ultimately we hope that this study gives researchers the tools they need to reduce the burden of coronary artery disease throughout the world," said Dr. Bjorkegren.
More information: A mechanistic framework for cardiometabolic and coronary artery disease, Nature Cardiovascular Research (2022). DOI: 10.1038/s44161-021-00009-1