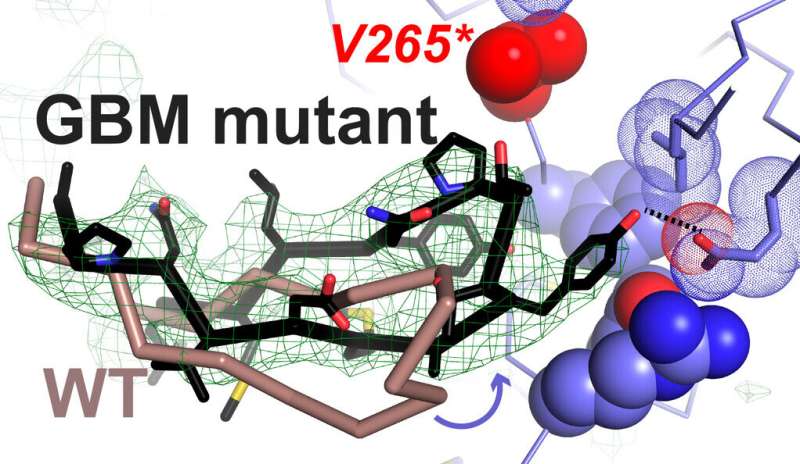
An electron density ‘omit map’ from X-ray crystallography showing a structural change in the EGFR dimerization site of the mutated receptor (indicated by green mesh), leading to enhanced EREG-driven dimerization. The mutated amino acid is colored red. Credit: Yale University
Researchers at the Yale Cancer Biology Institute have clarified the role of a recurring cancer-associated tumor mutation in glioblastoma multiforme (GBM), the most common and most aggressive type of malignant brain tumor. The findings were published today in Nature.
The epidermal growth factor receptor (EGFR) is frequently mutated in cancers and is an important and successful therapeutic target, especially in lung cancer. Yet, despite EGFR mutations occurring in approximately 24% of GBM cases, treatment with EGFR inhibitors has not been successful in this cancer.
In the new study, the research team showed that the most common extracellular mutations that occur in GBM prevent the EGFR from discriminating between the seven different growth factors that normally activate it. EGFR becomes activated when binding of a growth factor causes two receptors to come together into a dimer. Different growth factors normally stabilize EGFR dimers with distinct structures and activation kinetics, which signal differently to specify or bias cellular outcome. Some stop cell growth, while others (including a protein known as epidermal growth factor [EGF]) promote it. The team at the Yale Cancer Biology Institute found that mutated EGFRs seen in GBM react to all seven of the receptor's different ligands as if they are EGF, which induces the type of dimers that signal to promote cell proliferation.
"It was first thought that these mutations just activate the EGF receptor, much like the mutations seen in lung cancer, but our results show that they also change the type of signal given by the receptor," said Mark Lemmon, co-director of the Yale Cancer Biology Institute, deputy director of Yale Cancer Center, the David A. Sackler Professor of Pharmacology, and senior author of the study. "Our results provide new insight into how the EGF receptor differentiates one activating growth factor from another—and with these mutations, that ability to distinguish is lost."
In a collaborative effort across the Lemmon and Ferguson labs in the institute, Anatoly Kiyatkin, a research scientist at Yale, found that GBM mutations reduce the ability of EGFR to distinguish between EGF and epiregulin (EREG), another growth factor that binds to the EGF receptor in cellular assays. Graduate student Chun Hu, who is first author on the study, showed that the mutations allow EGFR to form strong dimers in response to EREG and other low-affinity ligands. Using X-ray crystallography, Hu worked with Carlos A. Leche, a Yale undergraduate, to show how two of the GBM mutations alter the process of receptor activation so that EREG drives the same type of signaling dimer normally seen with EGF.
Based on these findings, the researchers suggest an important role of altered ligand discrimination by EGFR in development of glioblastoma multiforme, with potential implications for therapeutic targeting in this and other cancers. "Our results showcase a nice combination of cellular work, solution biophysics, and crystallography—demonstrating how a tiny change in an amino acid side-chain can change the direction of the biology," said Lemmon.
GBM accounts for 48% of all primary malignant brain tumor diagnoses and more than 13,000 Americans are expected to be diagnosed with GBM this year. The five-year survival rate for patients with GBM is only 6.8%, and the average length of survival is estimated to be only 12 to 18 months.
More information: Mark Lemmon, Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias, Nature (2022). DOI: 10.1038/s41586-021-04393-3
Journal information: Nature
Provided by Yale University