Protective mutations in COVID-19
One way in which the body combats COVID-19 is by mutating the coronavirus, making it less harmful. This built-in protective mechanism in cells has a clear connection with decreased viral load in the body, a study from the University of Gothenburg shows.
Mutations are often associated with the emergence of virus variants that are more contagious and pathogenic than their predecessors. However, the current study shows that virus mutations often work in the opposite direction.
Virology researchers at the University's Sahlgrenska Academy have mapped mutation patterns in the SARS-CoV-2 coronavirus. The results, published in the journal PNAS, indicate that the body's natural enzyme ADAR1 (adenosine deaminases acting on RNA) impairs reproduction of SARS-CoV-2.
ADAR1, found inside the cells' protective membrane, can replace the nucleotides, which are the building blocks in the RNA of the virus. Nevertheless, how ADAR1 affects the coronavirus causing COVID-19 it has been unclear to date.
Mutations that benefit us
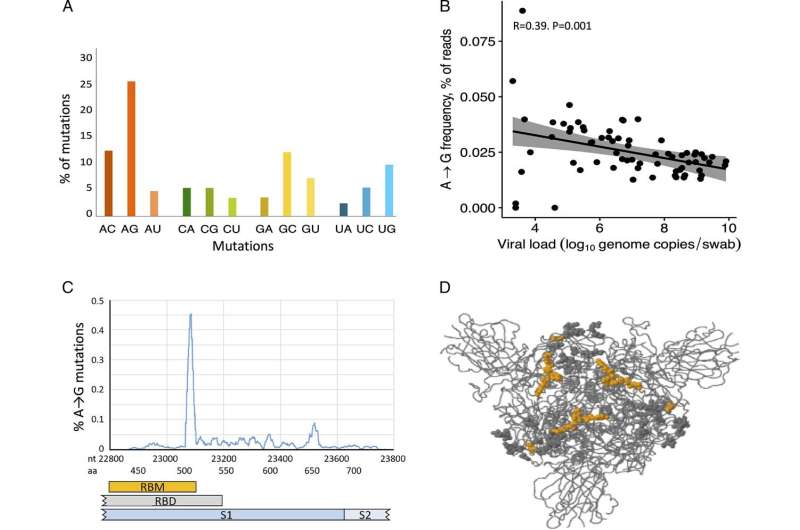
"Our study shows that there is an inverse relationship between the viral load (the measurable amount of virus in the body) and the extent to which ADAR1 has mutated the virus. We also found that ADAR1-induced mutations are the most common type of SARS-CoV-2 mutation," says Johan Ringlander, Ph.D. student in virology at Sahlgrenska Academy and the study's first author.
In particular, the scientists noted that individual patients are often infected with more than one variant of the virus. When mutations in relatively rare virus variants were investigated, it was found that a common mutation in which one nucleotide, guanosine (G), replaces adenosine (A) significantly worsened the reproductive ability of SARS-CoV-2. These mutations are caused by the enzyme ADAR1.
Analyses of more than 200,000 virus strains from patients who were ill with COVID-19 showed that mutations caused by ADAR1 were mainly circulating in summer 2020, when transmission and mortality rates were low in Europe. When transmission and mortality rates were higher, virus variants with ADAR1-induced mutations were uncommon, probably because they were outcompeted by more infectious virus strains.
Helps to clear away
"Our results clarify how the body's cells can generate mutated virus variants. Mutations can make a virus more infectious, but in most cases the mutations we've studied make the virus weaker; instead of spreading, it's removed from infected cells. These findings suggest that ADAR1 serves as a protective mechanism used by the body to limit viral infections," Ringlander says.
Michael Kann, Professor of Clinical Virology at Sahlgrenska Academy and chief physician at Sahlgrenska University Hospital, is the main author of the article.
"When SARS-CoV-2 multiplies in the airways, inflammation occurs. Its effects include activation of ADAR1, which in turn reduces the likelihood of the virus infecting other cells. We're currently investigating whether this protective mechanism may be important in other viral infections as well," Kann says.
More information: Johan Ringlander et al, Impact of ADAR-induced editing of minor viral RNA populations on replication and transmission of SARS-CoV-2, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2112663119