New methods to identify personalized drug treatments for breast cancer
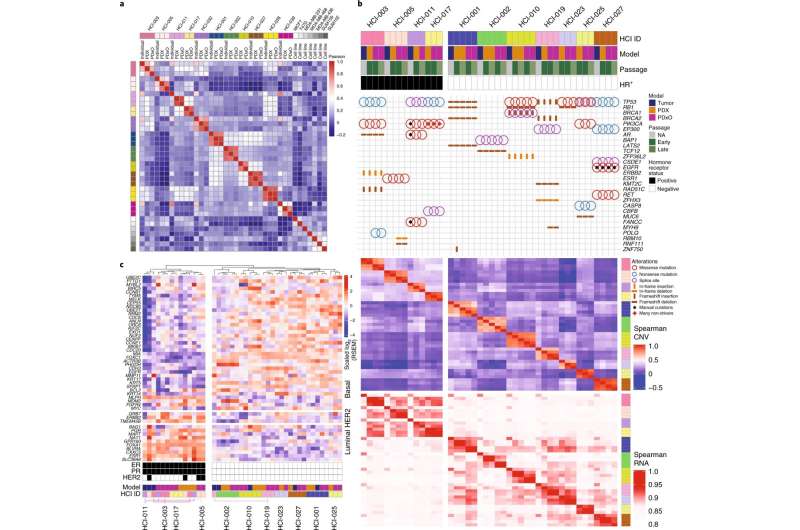
For years, researchers at Huntsman Cancer Institute at the University of Utah (U of U) have honed a process of developing breast cancer models using tumors donated by breast cancer patients, which they then implant into mice as a way to study the tumor's behavior.
Now, the research team reports a new, more efficient way to grow these tumors. In addition, they outline a process to test potential drugs to help prioritize clinical therapy choices based on unique tumor characteristics.
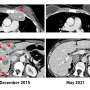
The study, published this week in the journal Nature Cancer, creates a way for researchers to narrow the number of drugs that might be effective in each tumor based on its unique characteristics and its behavior in the laboratory models of the cancer. Using this resource, the researchers uncovered experimental and Food and Drug Administration-approved drugs with high efficacy against the models. They extended this work to personalize therapy for a patient with metastatic breast cancer, which resulted in a complete response for the patient and a progression-free survival period more than three times longer than her previous therapies.
"We were able to utilize the data to prioritize therapy options for a patient," says Alana Welm, Ph.D., co-lead author, breast cancer researcher at Huntsman Cancer Institute, and professor of oncological sciences at the U of U. "While this therapy was unfortunately not curative, it led to regression of the patient's tumor and a longer survival period."
Welm says this unique bank of tumor models is critical to advancing research on aggressive breast cancers. "It is also, to our knowledge, the first time that such models have been used to influence the therapy choice of a breast cancer patient in a clinical trial setting."
The research team included a diverse group of clinicians, laboratory researchers, and technicians from Huntsman Cancer Institute at the University of Utah, Baylor College of Medicine, the Jackson Labs, the University of Connecticut, and the University of Pittsburgh. The team worked together to prioritize advancing research on samples most aligned with current challenges seen in the clinic.
A new clinical trial called FORESEE (NCT04450706) builds on the findings of this study. Led by Saundra Buys, MD, chief of the division of oncology at Huntsman Cancer Institute, the trial tests patient-derived tumor models to inform the treatment selection in metastatic breast cancer patients.
With a second trial in development, Welm says, "We will also use the models to predict recurrence for a subset of newly diagnosed breast cancer patients, and then attempt to personalize therapy for the metastatic stage of the disease when recurrence happens."
More information: Katrin P. Guillen et al, A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology, Nature Cancer (2022). DOI: 10.1038/s43018-022-00337-6