April 12, 2022 feature
Sequencing HIV proviruses from people on antiretroviral therapy using droplet-microfluidics
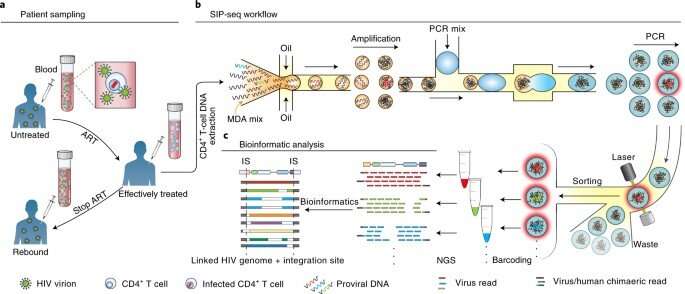
The infection of human immunology viruses (HIV) occurs by integrating its genome into infected cells to enter an inactive state of reversible latency that evades anti-retroviral therapy. The capacity to sequence such a provirus and adjacent host junctions in individual cells can highlight their mechanism of action and persistence in infected cells. However, this experiment is difficult to perform due to the 150-million-fold higher amount of background human DNA. In a new report now published in Nature Biomedical Engineering, Chen Sun and a research team in bioengineering and viral persistence and dynamics studies at the University of California, San Francisco, NIH and the Chan Zuckerberg Biohub, in the U.S., showed how full-length proviruses are connected to the HIV-host DNA junctions via high-throughput microfluidic assays, for droplet-based whole genome amplification of HIV DNA in its native context. The team conducted a polymerase chain reaction (PCR) to tag droplets containing proviruses to sequence and assay infected cells from people with HIV, receiving suppressing antiretroviral therapy at the time to detect and sequence paired proviral genomes and integration sites. The work sought to improve genetic analysis of persistent HIV-infected cell reservoirs.
The current state of HIV treatment and evolving therapeutic strategies
While approximately 37 million people worldwide are infected with human immunodeficiency virus (HIV), the cure remains unknown. The mechanism of infection of the virus imposes a central obstacle and also holds the key to curing the disease, as HIV integrates its genome into the genome of infected cells. While antiretroviral therapy (ART) can suppress virus replication to undetectable levels and prevent the progression of acquired immunodeficiency syndrome (AIDS), it lacks activity against cellular reservoirs. As a result, the therapy must be continued indefinitely to prevent virus rebound and disease progression. It is difficult to address HIV-associated stigma, due to the cost of lifelong ART administration, with toxic potential. It is therefore important to eradicate or suppress HIV-infected cellular reservoirs to obtain a functional cure. To examine the persistence of cellular reservoirs, scientists must conduct paired analysis of integration sites and full-length provirus sequences. By establishing a method that reliably and cost-effectively isolates and sequences rare pro-viruses, the researchers can comprehensively characterize HIV reservoirs. Sun et al developed such an approach to characterize proviruses and their cellular genomic context within HIV reservoirs using simultaneous integration site and provirus sequencing (SIP-Seq).
Beating the odds: A method to recover single provirus genomes
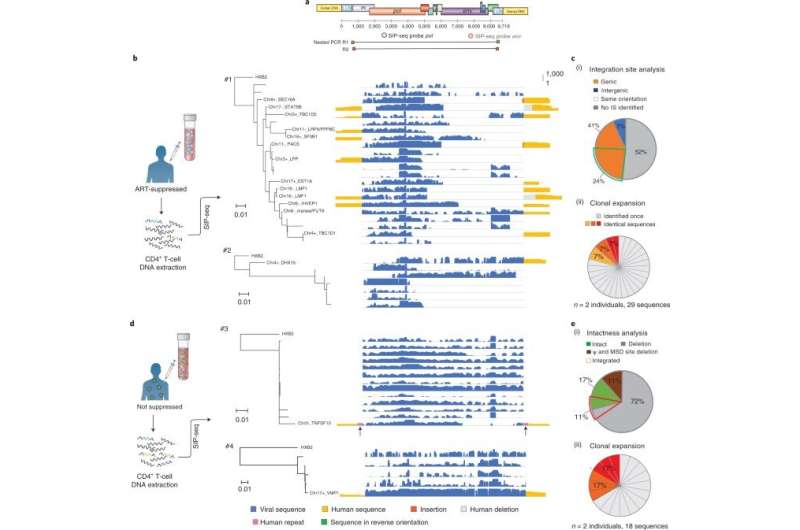
By using whole-genome sequencing amplification, Sun et al amplified the HIV genome in its native context within microfluidic droplets, then tagged the droplets containing proviruses for sequencing. The resulting method provided a full-length virus genome connected to its host-cell junctions via a single continuous assembly. The speed and efficiency of the method allowed the recovery of single provirus genomes in a 150-million-fold higher background of DNA. The team used SIP-Seq (simultaneous integration site and provirus sequencing) to profile the provirus population in a variety of ART-related individuals to understand the latent HIV reservoir. While the team focused on HIV, this approach is applicable to diverse viruses that invade the host's genome, to provide a general platform to characterize genetics of a variety of infections.
The experiments
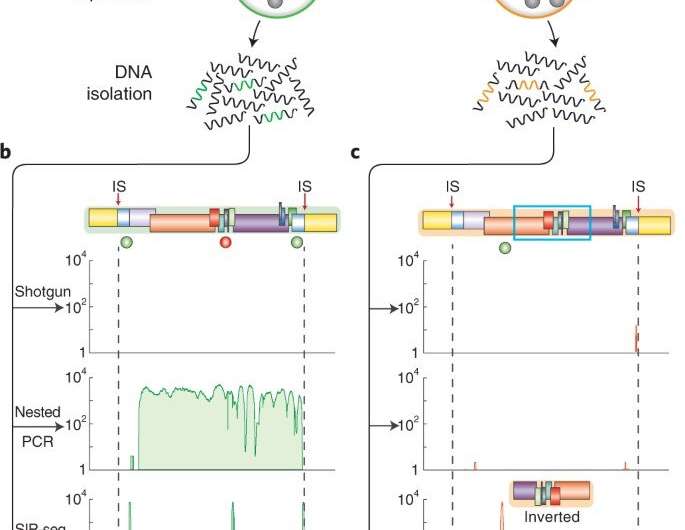
Sun et al aimed to recover all DNA fragments containing HIV genomes from a population of millions of CD4+ T cells, to sequence the molecules individually, and their immediately adjacent junctions. The team extracted large DNA fragments from cells encapsulated in micro-droplets to isolate the integrated HIV genomes. Using multiplexed displacement amplification (MDA), they non-specifically amplified each genomic fragment to obtain sufficient single proviruses for sequencing. They then followed this by isolating individual droplets with HIV proviruses, where each droplet was barcoded and sequenced. The virologists then mapped readouts from each droplet to an HIV reference genome to identify integration sites to facilitate clear-cut information on the complexity of the virus genome, and their integration sites—crucial to assess the cellular reservoir.
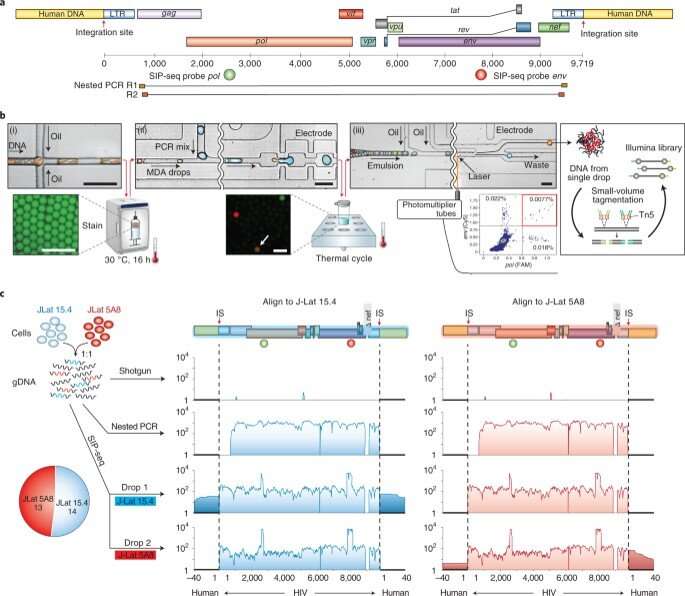
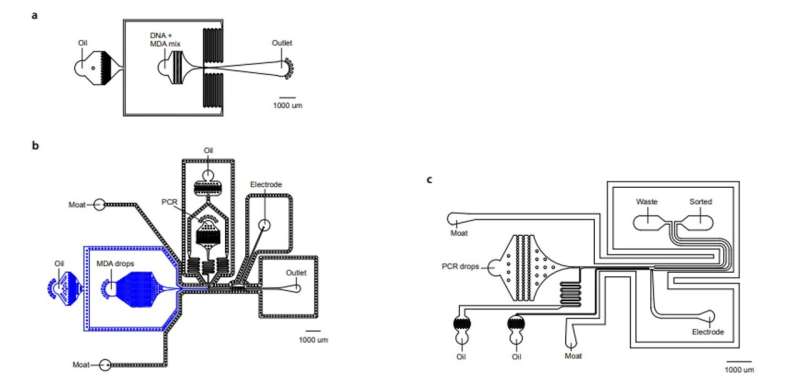
Detecting proviruses with SIP-Seq and regulating microfluidics
To increase the specificity of full-length proviruses and prevent random DNA cleavage, Sun et al employed a dual specific multiplexed Taqman PCR, to target regions on the HIV genome. During the experiments, they yielded a specific volume of DNA to sequence and confirm HIV provirus recovery. The microfluidics of the SIP-Seq setup contained three devices, including a droplet encapsulator, a merger and a sorter. The scientists loaded the human genomic DNA fragments with multiple displacement amplification (MDA) reagents to encapsulate up to 10 billion DNA fragments, approximating 75 kbp in length, in 20 million separate droplets, within 15 minutes. Each droplet contained 500 distinct fragments and Sun et al incubated them at 30 degrees Celsius, for non-specific amplification, prior to merging with Taqman PCR reagents. After repeating the steps for all detected HIV genomes, the team prepared the tubes for sequencing.
Analyzing HIV proviruses and their genomic landscape in ART-treated persons
Sun et al next validated the SIP-Seq on HIV cell lines and analyzed HIV in persons treated with ART. During the experiments, they prepared cells by plating, resting CD4+ T cells from an ART-treated person, followed by stimulation and in vitro culture for proliferation via three methods; shotgun, nested PCR, and SIP-Seq with Taqman probes. Both SIP-Seq and nested PCR yielded excellent results, the team specifically opted for SIP-Seq to characterize the full genetic diversity of infected cells in vivo. They followed this work by characterizing the genomic landscape of HIV proviruses, which they directly isolated from CD4+ T cells of infected persons. The outcomes revealed three clonal lineages to support clonal origin relative to previous studies of the same individuals.
Outlook
The research indicated how the methods enabled Chen Sun and colleagues to study HIV proviruses more efficiently than with existing PCR methods. The outcomes provided a comprehensive analysis of the HIV genetic landscape in vivo by characterizing the latent reservoir to highlight its role in HIV persistence. Existing methods were challenging due to the rarity and lack of distinct surface markers to identify latently infected cells. Using the described SIP-Seq (simultaneous integration site and provirus sequencing) method, Sun et al provided a fast, cost-effective and scalable process, which they applied to people receiving antiretroviral therapy (ART), to highlight the presence in latent reservoirs of clonally expanded cells. The strategy is applicable across HIV-infections, to other viral diseases with integrated states in their life cycle.
More information: Chen Sun et al, Droplet-microfluidics-assisted sequencing of HIV proviruses and their integration sites in cells from people on antiretroviral therapy, Nature Biomedical Engineering (2022). DOI: 10.1038/s41551-022-00864-8
Katherine M. Bruner et al, A quantitative approach for measuring the reservoir of latent HIV-1 proviruses, Nature (2019). DOI: 10.1038/s41586-019-0898-8
© 2022 Science X Network