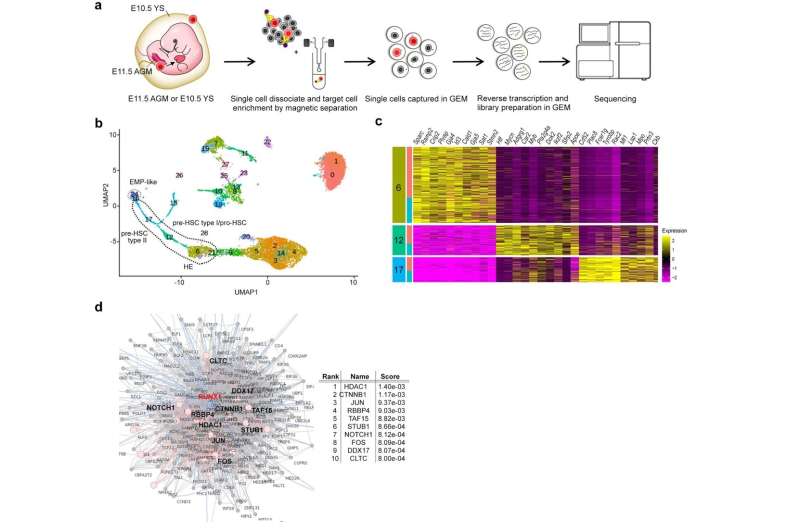
Figure 1. a: Experimental schematic. Cells are isolated from the fetal yolk sac and AGM and their transcripts are sequenced by next-generation sequencing. b: Cell groups from single-cell analysis of AGM progenitor cells are shown. c: Heatmap of top 10 enriched genes in the three cell groups (6, 12, 17) in b. d: Gene enrichment analysis based on protein-protein inter-network of RUNX1, a transcription factor essential for the endothelial-to-hematopoietic transition (EHT), in which vascular endothelial cells give rise to hematopoietic stem cells. Credit: Figure derived from the paper published in Molecular Psychiatry (2022) (DOI:10.1038/s41380-022-01566-y)
An international research group led by Professor Toru Takumi (Senior Visiting Scientist, RIKEN Center for Biosystems Dynamics Research) and Researcher Chia-wen Lin at Kobe University Graduate School of Medicine has shown that idiopathic autism is caused by epigenetic abnormalities in hematopoietic cells during fetal development, which results in immune dysregulation in the brain and gut. The results of the study revealed that in autism, there are immune abnormalities that can be seen in the brain and gut. The further classification of the pathophysiology of autism could lead to the creation of new treatment strategies for autism and other neurodevelopmental disorders. The results of this research will be published in Molecular Psychiatry on Monday May 2, 2022.
Research background
Autism spectrum disorder is a developmental neurological disorder that remains largely unexplored despite the rapidly increasing number of patients. Immune abnormalities, now considered the cause of many diseases, also play an important role in the development of autism. Brain inflammation and disturbances of the peripheral immune system are frequently observed in autistic patients. Furthermore, immune abnormalities are accompanied by abnormalities in the intestinal microbiota, which is also thought to be involved in the pathogenesis of the disease via the brain-gut axis. However, the essential mechanisms behind these immune abnormalities have yet to be elucidated.
Given the critical developmental stages of immune insults and the extensive involvement of the immune system in the development of autism, the research team hypothesized that a common etiology underlies the widespread immune dysregulation and originates in different types of progenitor cells. The analysis focused on the hematopoietic cells from which immune cells are derived, as well as on the yolk sac (YS) and the aorta-gonad-mesonephros (AGM), which are involved in hematopoiesis during the fetal stage. These results seek a common ancestor of inflammation in the brain and abnormalities in the peripheral immune system. In this study, BTBR mice were used as an idiopathic model for autism.
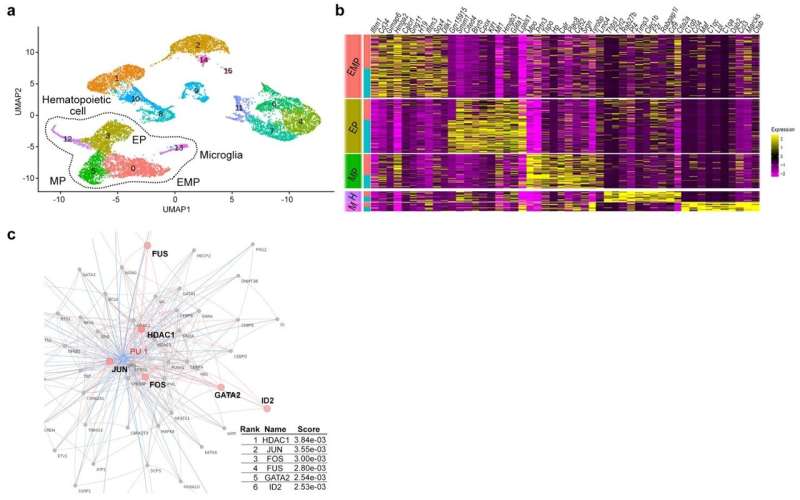
Figure 2. a: Cell groups from single-cell analysis of yolk sacs. b: Heat map of the top 10 genes enriched in the five cell groups in a. c: Protein-protein interaction of PU.1, a transcription factor important for microglial development. Network-based gene enrichment analysis. Credit: Figure derived from the paper published in Molecular Psychiatry (2022) (DOI:10.1038/s41380-022-01566-y)
Research findings
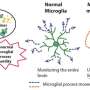
Single-cell RNA sequencing (sc-RNA seq) of BTBR mice traced the origin of immune abnormalities back to the embryonic stages of the yolk sac and aorta-gonad-mesonephros (AGM) and identified where macrophages (microglia) and peripheral immune cells differentiate. Definitive hematopoiesis in YS and AGM single-cell level analysis successfully identified pathological mechanisms at the molecular level within rare progenitor cells in the early stages of development. Namely, we found a common mechanism of transcriptional regulation through HDAC1, a histone deacetylase, underlying these pathologies (Figures 1 and 2).
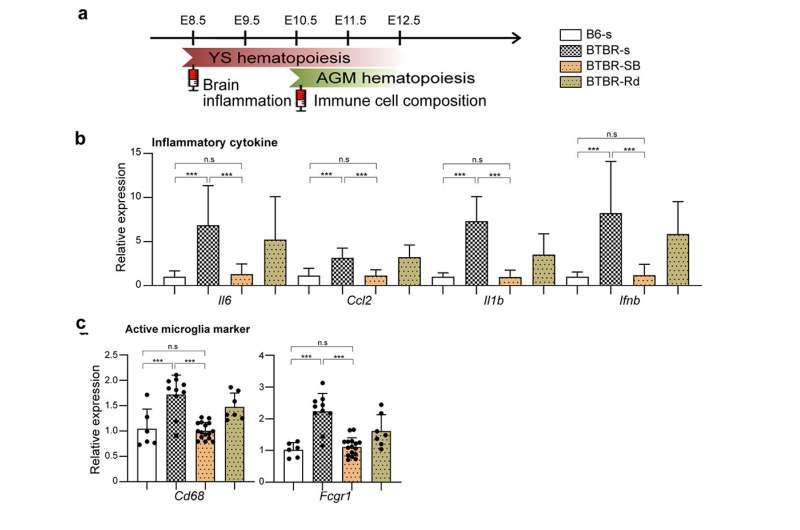
We have also shown that manipulating epigenetic mechanisms during specific developmental stages can restore immune abnormalities in the brain and peripheral tissues. Namely, we identified histone deacetylase HDAC1 as a common mechanism. Administrating inhibitors of this histone (sodium butyrate or Romidepsin) during the fetal stage in BTBR mice suppressed elevated inflammatory cytokines and microglial activation (Figure 3).
Figure 3. a: Inhibitors, sodium butyrate (SB) or Romidepsin (Rd) administered at embryonic 8.5 and 10.5 days, the beginning of primary hematopoiesis in the yolk sac and AGM, respectively, were compared to the control saline (s) group. b: In BTBR brain inflammatory cytokines (Il6, Ccl2, Il1b, Ifnb) were suppressed. c: Markers of activated microglia (Cd68, Fcgr1) were suppressed. Credit: Figure derived from the paper published in Molecular Psychiatry (2022) (DOI:10.1038/s41380-022-01566-y)
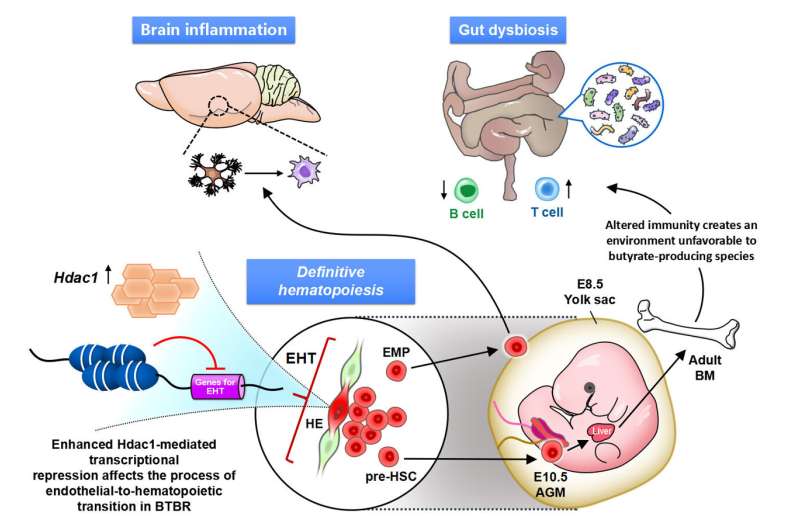
We further demonstrated that dysregulated immunity can determine gut dysbiosis of specific profiles in autistic model mice, which make the potential biomarkers of Treg and gut dysbiosis a means to categorize the immune-dysregulated ASD subtype. From the above, it is clear that the abnormalities in the brain and peripheral organs (such as the intestines) seen in autism are caused by epigenetic abnormalities in the hematopoietic stem cell lineage, the ancestor of immune cells (Figure 4).
Figure 4. Transcriptional repression by promoting histone deacetylase 1 (HDAC1) affects the process from vascular endothelial cells to the production of hematopoietic stem cells (EHT). Erythroid/myeloid progenitor cells (EMP) in the yolk sac (Yolk sac) activate microglia and affect inflammation in the brain; pre-HSC in AGM differentiate in the bone marrow (BM); and the altered immune system affects the gut microbiota. Credit: Figure derived from the paper published in Molecular Psychiatry (2022) (DOI:10.1038/s41380-022-01566-y)
Perspectives
Our findings not only provide the missing piece to solve the long-time puzzle of systemic immune dysregulation in autism, but also hint the role of epigenetic disturbance as common etiology among different autism models of environmental risk factors. Furthermore, to develop precision medicine for ASD in the future, ASD subtyping according to the pathogenesis mechanism is a key first step to resolve the heterogeneity of ASD and to open up a new avenue for ASD treatment.
More information: Chia-Wen Lin et al, A common epigenetic mechanism across different cellular origins underlies systemic immune dysregulation in an idiopathic autism mouse model, Molecular Psychiatry (2022). DOI: 10.1038/s41380-022-01566-y
Provided by Kobe University