Dog coronavirus jumps to humans, with a protein shift
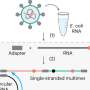
![Positive selection, unique amino acid changes and GARD partitions mapped to a CCoV-HuPn-2018 spike domain map [3]. S1 and S2 of the protein are highlighted and further subdivided into functional subunits and subdomains. Blue dots represent sites under positive selection in CCoV-HuPn-2018 as identified by MEME and/or FEL; red dots represent sites that are unique in CCoV-HuPn-2018 but are not under positive selection; yellow dots are nonsynonymous changes between CCoV-HuPn-2018 and HuCCoV_Z19Haiti. Text labels accompany each subdomain/functional unit: SP, signal peptide; 0 domain; A domain; B, includes RBD-Receptor-Binding Domain; C; D; UH, upstream helix; FP, fusion peptide; HR1, heptad-repeat 1; CH, central helix; BH, β-hairpin; CD: connector domain; HR2, heptad-repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. The horizontal magenta bar represents the experimentally evaluated region for sialic acid binding in TGEV [11,37]. The solid vertical black lines represent the breakpoints of the GARD identified non-recombinant fragments and are labeled numerically. The vertical dashed line represents the 3′ end of alignment set I and the onset of FCoV2 sequence homology (alignment set II). Credit: Viruses (2022). DOI: 10.3390/v14050853 Dog coronavirus jumps to humans, with a protein shift](https://scx1.b-cdn.net/csz/news/800a/2022/dog-coronavirus-jumps.jpg)
Cornell University researchers have identified a shift that occurs in canine coronavirus that may provide clues as to how it transmits from animals to humans.
A new canine coronavirus was first identified in two Malaysian human patients who developed pneumonia in 2017-18. A group of other scientists isolated the canine coronavirus, sequenced it, and published their findings in 2021.
Now, a team led by researchers from Cornell and Temple University has identified a pattern that occurs in a terminus of the canine coronavirus spike protein—the area of the virus that facilitates entry into a host cell. This pattern shows the virus shifts from infecting both the intestines and respiratory system of the animal host to infecting only the respiratory system in a human host.
The researchers identified a change in the terminus—known as the N terminus—a region of the molecule with alterations also detected in another coronavirus, which jumped from bats to humans, where it causes a common cold.
"This study identifies some of the molecular mechanisms underlying a host shift from dog coronavirus to a new human host, that may also be important in the circulation of a new human coronavirus that we previously didn't know about," said Michael Stanhope, professor of public and ecosystem health at Cornell. First author, Jordan Zehr, is a doctoral student at Temple University. The paper was published in the journal Viruses.
In the study, the researchers used state-of-the-art molecular evolution tools to assess how pressures from natural selection may have influenced the canine coronavirus' evolution.
The same variant of canine coronavirus found in Malaysia was also reported in 2021 in a few people in Haiti, who also had respiratory illness.
Stanhope believes more study is needed to understand if the viral shifts and jumps to humans occurred spontaneously in different parts of the world or if this coronavirus has been circulating for perhaps many decades in the human population without detection.
More information: Jordan D. Zehr et al, Recent Zoonotic Spillover and Tropism Shift of a Canine Coronavirus Is Associated with Relaxed Selection and Putative Loss of Function in NTD Subdomain of Spike Protein, Viruses (2022). DOI: 10.3390/v14050853