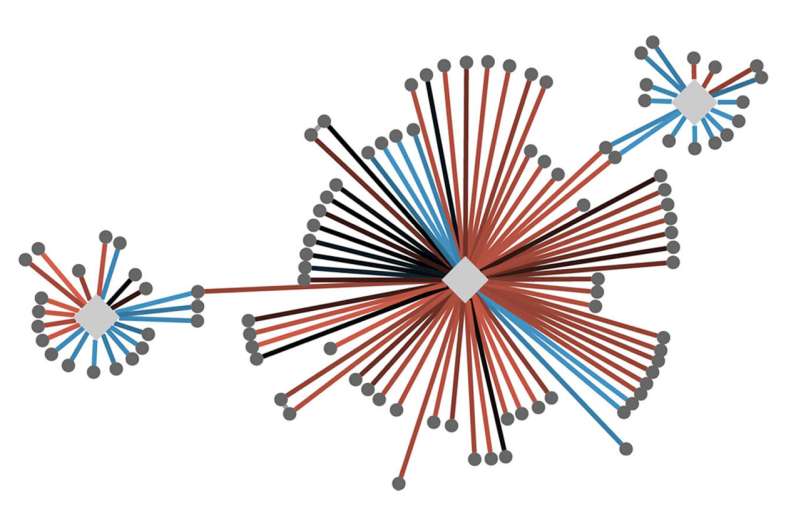
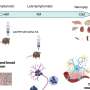
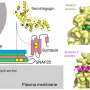
A generic version of a cellular protein interaction map. The diamond shapes represent proteins of interest, which have been experimentally characterized to participate in certain “protein complexes” in the cell that come together to facilitate a particular cell function. The lines emanating outward show the physical interactions between proteins of interest (diamonds) and their interacting partners (circles). These cellular maps help researchers see how complexes can be disrupted when proteins are mutated, and to consider how potential therapies could affect the network. Credit: Krogan Lab
Breast cancer, COVID-19, and autism may seem unrelated, but they share some surprising connections. Some of the same genes that are mutated in breast cancer also get hijacked by COVID-19, and some other genes mutated in cancer are also implicated in autism.
Commonalities like these have led Nevan Krogan, Ph.D., director of UCSF's Quantitative Biosciences Institute, to examine in detail the effects of a handful of genes that seem to play an outsize role in a wide array of diseases.
Those effects rely on proteins, for which genes are the blueprints. When a gene is mutated, so is its protein.
"Our genome is relatively static, but proteins aren't," said Krogan. "They're constantly interacting with other proteins in different contexts that change over time."
Many conditions involve dozens of mutations, he added. Seeing the full landscape of a person's disease means piecing together how each of those mutated proteins contributes to it.
More than a decade ago, Krogan began employing sophisticated quantitative approaches to create "cell maps" that compare thousands of these protein-protein interactions, or PPIs, in healthy and diseased cells across a range of mutations in cancer, autism, and infectious disease.
He believes that zeroing in on these PPIs can elucidate how mutations disrupt cell functions and uncover entry points for safer and more effective treatments.
Already, collaborations between Krogan and researchers in the U.S. and around the world have revealed how mutations in different genes sometimes bungle the same cellular pathways, illuminating connections between diseases that may look quite different at the genetic level.
In other cases, the same gene is implicated in more than one disease: a mutation at point A may contribute to cancer, while a mutation at point B may create a predisposition to a psychiatric disorder.
"We're finding the Achilles' heels of the genome," Krogan said. "By going beyond DNA and looking at these networks of protein interaction, we're able to connect dots that we didn't even know existed before."
Nevan Krogan, Ph.D. Credit: Noah Berger
Mapping the network
To find those dots and draw the lines between them, Krogan, along with his collaborators, use his cell maps to see exactly how a specific mutation in a particular gene translates into changes in protein interactions.
A gene called PIK3CA, for example, is involved in a sizable percentage of cancers, as well as autism and other brain disorders. There are hundreds of known mutations in PIK3CA, each of which has a specific effect on the protein machinery.
Krogan has catalogued not only how each of these mutations leads to disease but also how PIK3CA's various pathways play out in healthy cells, allowing him to identify the intersection where each of these mutations throws the cell's protein interactions off track.
Achieving this granular approach involves overlaying large sets of data and finding patterns that pinpoint the molecular moment when a cellular process goes awry. Krogan's teams use mass spectrometry to weigh the protein molecules and combines it with other methods that assess the protein's structure. Advanced computational techniques are needed to crunch the enormous amount of data involved.
These maps can help provide a prognosis based on proteins resulting from the mutations found in a particular patient's genes; help clinicians choose one treatment over another; and reveal where a drug might be able to stop a disease without interfering with other healthy cell functions.
A new view of disease
While some researchers have studied PPIs associated with individual gene mutations, Krogan has spent his career investigating them on a massive scale. "There's great value in looking at the big picture," he said. "It makes these analyses exponentially more powerful."
Krogan likens the protein maps to a computer-generated geographic map. You can zoom out to see a large area, then zoom in to see local detail, then zoom back out again to put that detail into context.
Being able to see those varying levels of detail can potentially help researchers identify FDA-approved drugs that could be tested for unexpected applications, said Krogan. "These cell maps are an entirely new way of looking at disease and drug discovery."
Ultimately, Krogan's goal is to enable researchers to apply artificial intelligence to these maps, so they can predict a patient's prognosis and the best combination of drugs to treat them.
"Once we understand this underlying biology, attacking the disease becomes so much more straightforward," Krogan said. "We're perfectly positioned to build this bridge from the genome to the clinic for a whole range of disorders."
"We're at the cusp of such great things."
Provided by University of California, San Francisco