A refined bioinformatic analysis can estimate gene copy-number variations from epigenetic data
A team led by Dr. Manel Esteller, Director of the Josep Carreras Leukemia Research Institute, has improved the computational identification of potentially druggable gene amplifications in tumors, from epigenetic data. By using this new tool, scientists can have reliable information on the molecular data of a particular tumor, both at the genetics and epigenetics level, even from scarce samples.
Cancer cells exhibit a plethora of genetic and epigenetic alterations that are both the cause and the consequence of their deep dysregulation. Epigenetic abnormalities impair the proper function of genes, either by increasing or decreasing their activity, while genetic alterations often imply mutations or gains and losses of entire genes or chromosome fragments. The latter, named Somatic Copy Number Alterations (SCNAs) have been associated with anticancer drug response: the higher the copy number amplification of a target gene, the higher its sensitivity to the targeted treatment.
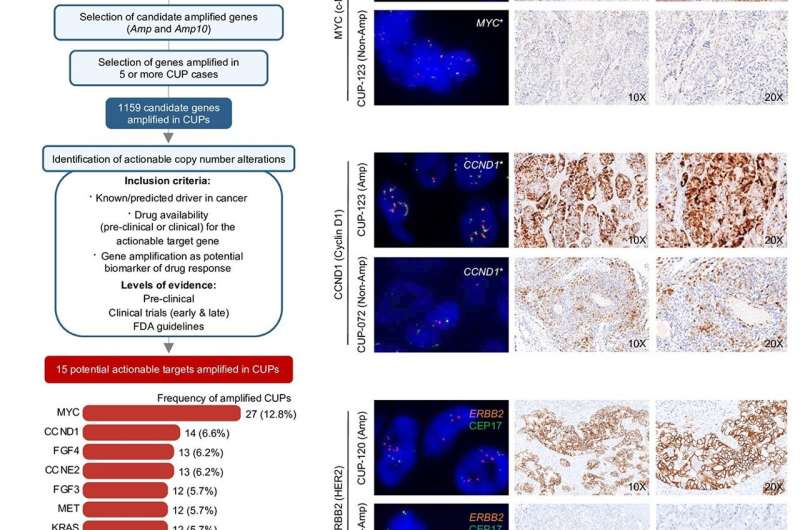
Genetic and epigenetic profiling of a tumor can help define its prognosis and treatment options. However, it takes a large sample to do both analysis when the amount of sample is usually limited. The researchers' team at the Josep Carreras Leukemia Research Institute have improved a previous tool (conumee) to develop the new conumee-Kcn, which is capable to improve the characterization of SCNAs up to a 20%, and distinguish between low gains, moderate amplifications and high amplifications.
The new methodology, published at the scientific journal Briefings in Bioinformatics, uses dynamic thresholding and tumor purity estimates over DNA-methylation array data to infer SCNAs with high accuracy, reaching a 100% specificity and 93.3% sensitivity in the preliminary experimental tests, using the gold standard fluorescence in situ hybridization (FISH) assay. Hence, with the same information used to profile the epigenetics signature of a tumor, they can estimate genetic copy number variations, a highly relevant clinical feature; two birds with one stone.
Though this estimation is not as good as when using the standard SNP arrays, a methodology specifically designed to SCNA analysis, it offers valuable information when facing limited samples and resampling is not possible or advised, like in lung cancers.
After the benchmarking, the researchers tested the reliability of conumee-Kcn on stored samples from Cancers of Unknown Primary (CUPs), a type of highly aggressive metastatic tumors with poor prognosis due to its unknown origin. Dr. Verónica Dávalos, lead co-author of the research, states that up to 15 potentially druggable targets were identified in those samples, opening the door to new treatment opportunities in the future for this dismal tumor type.
The capacity to treat CUPs, and any other cancer type, only by their genetic and epigenetic alterations, regardless of their origin, is a new approach to fight cancer in a tissue-agnostic manner. On this regard, the Cancer Epigenetics group at the Josep Carreras Leukemia Research Institute is deepening its knowledge on the molecular features of CUPs to bring new treatment options to the clinician's arsenal.
More information: Pedro Blecua et al, Refinement of computational identification of somatic copy number alterations using DNA methylation microarrays illustrated in cancers of unknown primary, Briefings in Bioinformatics (2022). DOI: 10.1093/bib/bbac161