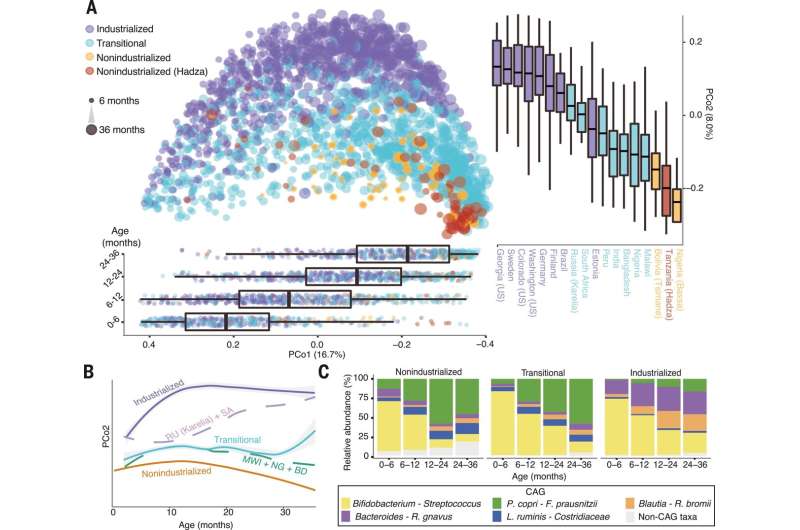
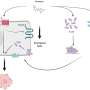
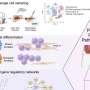
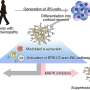
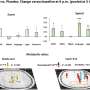
Age and lifestyle are associated with infant microbiome composition. (A) Unweighted UniFrac dissimilarity Principal Coordinates Analysis (PCoA) (top left panel) of 1900 fecal samples from infants (<3 years old) across 18 populations based on amplicon sequence variant abundance. Point color indicates lifestyle and point size is proportional to age in months. Boxplots show the distribution of indicated age groups along PCo1 (bottom) and cohorts along PCo2 (right). (B) PCo2 versus sample age for the three lifestyle categories (solid lines) and specific indicated subpopulations (dashed lines). The purple dashed line includes Russia (Karelia) and South Africa [RU (Karelia) + SA] and the green dashed line includes Malawi, Nigeria (Urban), and Bangladesh (MWI + NG + BD). The middle transitional line (blue) contains all transitional samples. Lines are the smoothed conditional mean of PCo2 loadings (loess fit). (C) Relative abundance of CAGs by age group and lifestyle. Taxa in annotation are the most abundant taxa in a CAG. Credit: Science (2022). DOI: 10.1126/science.abj2972
A team of researchers from Stanford University, Chan Zuckerberg Biohub, the University of California, Berkeley, and New York University Abu Dhabi has found that the gut microbiome of infants living in a hunter-gatherer society in Tanzania is markedly different from the gut microbiome of infants living in modern urban areas. In a study published in the journal Science, the group conducted ribosomal RNA sequencing on more than 100 stool samples obtained from Hadza infants and compared them to similar data stored in public databases.
Noting that most biome gut sequencing research has been conducted on people living in urban areas, the researchers wondered about the gut biomes of people living in remote, non-urban settings. Noting also that some prior research has shown that people living in such areas tend to have a more diverse gut biome than people living in urban areas, they wondered about the gut biomes of infants in such places.
The team collected stool samples from dozens of Hadza infants living in Tanzania, along with stool samples collected from 23 of the infant's mothers. They then conducted ribosomal RNA sequencing on all of the samples to determine the kinds of bacteria in their guts that make up the biome. They compared the diversity of the gut microbiomes in the infants in Tanzania with those in the guts of infants living in modern urban areas around the world. They found that the Hadza infants had more diversity in their guts after approximately six months than did infants living in urban areas. They also found that approximately 20% of the bacteria types found in the Hadza infants' microbiomes had not been previously documented.
The researchers also found that the differences in the gut microbiome could be traced back to their mothers along with some influences from the local environment. They also suggest the main reason for the differences in the gut microbiome appeared to be related to lifestyle rather than geography. And they also speculate on the possibility of a link between a less diverse gut microbiome in urban areas and diseases that are more common in the industrialized world—such as those related to inflammation.
More information: Matthew R. Olm et al, Robust variation in infant gut microbiome assembly across a spectrum of lifestyles, Science (2022). DOI: 10.1126/science.abj2972
Journal information: Science
© 2022 Science X Network