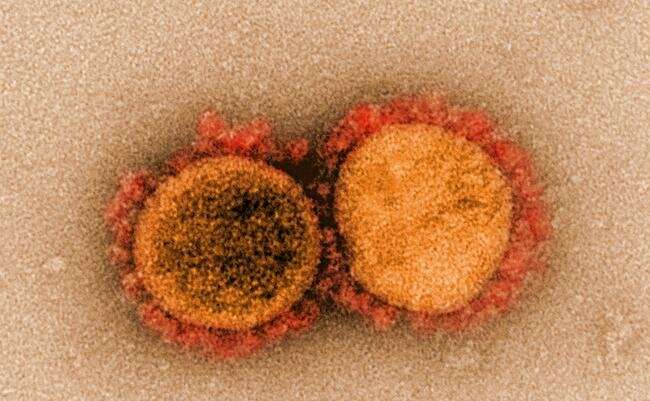
Transmission electron micrograph of SARS-CoV-2 virus particles, isolated from a patient. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
With the end of the pandemic seemingly nowhere in sight, scientists are still very focused on finding new or alternative drugs to treat and stop the spread of COVID-19. In a first-of-its-kind study, researchers at the University of New Hampshire have found that using an already existing drug compound in a new way, known as drug repurposing, could be successful in blocking the activity of a key enzyme of the coronavirus, or SARS-CoV-2, which causes COVID-19.
"The goal was to slow or prevent the spread of the virus by using a strategic therapeutic that could possibly disrupt key steps in the viral life cycle at the molecular level, like the first contact with a healthy cell or the first step in replicating within an infected cell," said Harish Vashisth, associate professor of chemical engineering.
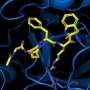
In their study, recently published in the journal Proteins: Structure, Function, and Bioinformatics, researchers set out to target a key enzyme responsible for COVID-19, called the main protease enzyme Mpro, which has become a primary target of intense research and therapeutic development because it is essential for the virus to replicate. In this case, they explored the inhibiting properties of a derivative of the potent chemical compound known as Thiadiazolidinones, or TDZD, which are already being studied as a potential treatment for neurological disorders like Parkinson's Disease. Researchers used a specific TDZD compound, known as CCG-50014, to target Mpro which acts like a molecular scissor by cutting up long chains of polypeptide proteins of the virus into smaller component proteins. These smaller segments can fold and mature to form new virus particles. Using molecular dynamics simulations combined with laboratory experiments, the researchers determined that TDZD compound was able to inhibit the Mpro enzyme.
"Coronaviruses, like COVID-19, are a notorious group of infectious agents that include a large class of viruses with RNA genomes, similar to the human DNA genome, that depend on well-organized protein structures crucial for viral growth and replication," said Vashisth. "These viruses can develop rapid defenses at the cellular level by orchestrating these layers, or folding mechanisms, in viral proteins so the key is to find a way to shut them down."
RNA viruses are known for causing seasonal epidemics, like influenza, and can appear as novel virus strains with high fatality rates (COVID-19, SARS, Zika and Ebola). Researchers say the need for an alternative drug development pipeline, instead of the intensive process of introducing new drugs to market, is illustrated by the high infection rate of COVID-19 (compared to previous coronaviruses) and is important for a long-term effective response to new and reoccurring outbreaks.
More information: Jacob Andrzejczyk et al, Molecular interactions and inhibition of the SARS‐CoV‐2 main protease by a thiadiazolidinone derivative, Proteins: Structure, Function, and Bioinformatics (2022). DOI: 10.1002/prot.26385
Provided by University of New Hampshire