Researchers uncover a novel host cell pathway hijacked during COVID-19 infection
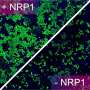
An international team of scientists, led by the University of Bristol, has been investigating how the SARS-CoV-2 virus, the coronavirus responsible for the COVID-19 pandemic, manipulates host proteins to penetrate into human cells. After identifying Neuropilin-1 (NRP1) as a host factor for SARS-CoV-2 infection, new findings published in the journal of the Proceedings of the National Academy of Sciences describe how the coronavirus subverts a host cell pathway in order to infect human cells.
SARS-CoV-2 continues to have a major impact on communities and industries around the world. In an attempt to find innovative strategies to block SARS-CoV-2 infection, the team previously identified NRP1 as an important receptor at the surface of cells that is hijacked by SARS-CoV-2 to enhance infection.
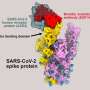
NRP1 is a dynamic receptor that senses the microscopic cellular environment through the recognition of proteins containing specific neuropilin-binding sequences, called ligands. By mimicking this neuropilin-binding sequence, SARS-CoV-2 is able to subvert this receptor to enhance its entry and infection of human cells.
In this new study, the group led by Bristol's Faculty of Life Sciences, Professor Peter Cullen from the School of Biochemistry and Drs Boris Simonetti, Senior Researcher and James Daly, Research Associate, in the Cullen lab, has now identified NRP1 and its ligands are transported within the host cell by a protein complex, called ESCPE-1. The protein complex captures NRP1 and regulates its transport between compartments inside the cell.
The function of this pathway is still not completely clear, but the team found that using gene editing to remove ESCPE-1 from human cells effectively blocked SARS-CoV-2 infection by around 50%, suggesting that this process is beneficially hijacked by the virus during the infection process.
Pete, Boris and James, explained that "this study represents an advance in the understanding of the pandemic coronavirus, and how it subverts host biology in order to infect cells. The identification of this pathway used by SARS-CoV-2 opens avenues for designing therapeutic interventions that can prevent ESCPE-1 and NRP1 from associating with the Spike protein to reduce infection."
More information: Boris Simonetti et al, ESCPE-1 mediates retrograde endosomal sorting of the SARS-CoV-2 host factor Neuropilin-1, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2201980119