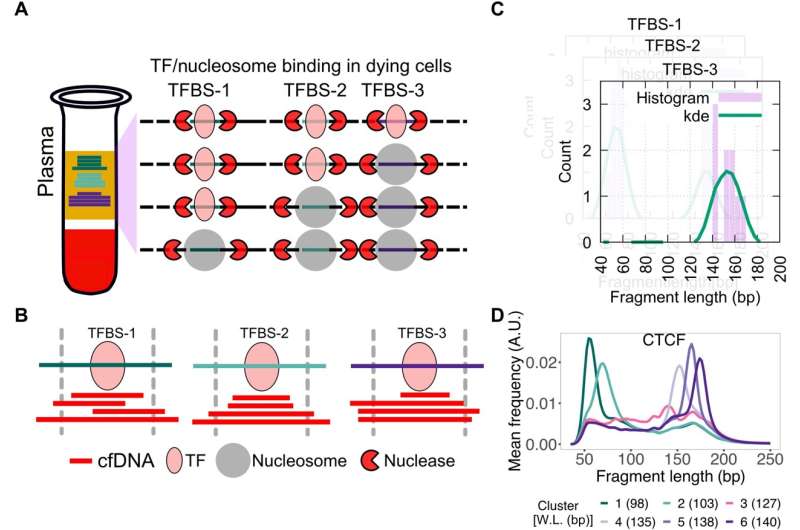
Schematic for identifying subset of binding sites with TF footprints. (A) When TFs or nucleosomes are bound at TFBSs, they protect different lengths of DNA from nucleases in dying cells in the human body. (B) When sequenced cfDNA fragments are mapped to TFBSs ± 50 bp, varying numbers of short and long cfDNA fragments are found at the three TFBSs shown in (A). (C) cfDNA fragment length distribution is estimated at each TFBS (purple bars) and smoothed using kernel density estimation (kde) (green line). (D) K-means clustering is performed on smoothed length distribution to group TFBSs with similar cfDNA fragment length distribution. Here, smoothened length distributions of clusters of CTCF TFBS are shown. Weighted length (W.L.) for each CTCF length cluster is shown in parentheses. A.U., arbitrary units. Credit: Science Advances DOI: 10.1126/sciadv.abm4358
Researchers at the University of Colorado Anschutz Medical Campus have discovered how to extract critical information about breast cancer tumors and disease progression by analyzing blood plasma rather than using more invasive tissue biopsies.
"This is simply a blood draw," said the study's senior co-author Peter Kabos, MD, associate professor of medicine in the medical oncology division at the University of Colorado School of Medicine and CU Cancer Center member. "This allows us to look under the surface to see the defining characteristics of the disease. The advantage is that we don't need to do repeated tissue biopsies."
The study was published today in the journal Science Advances.
Researchers and clinicians analyze plasma to define gene mutations in cancer. The DNA found in plasma contains much more information.
"We just need to know where to look," said Kabos.
The study found that plasma cell-free DNA (cfDNA) contains high resolution, genome-wide binding estrogen receptors (ER) and FOXA1 profiles for breast cancer. FOXA1 is a gene associated with breast cancer.
"We can obtain the same molecular information we get from tissue biopsies directly from the blood," said the study's other senior co-author Srinivas Ramachandran, Ph.D., assistant professor of biochemistry and molecular genetics at the CU School of Medicine. "If we can extract this kind of information without doing a biopsy, we could provide more information for treatment decisions."
Dying cells in the human body release their content into the bloodstream, the researchers said. When cancer is present, it also releases fragments of cfDNA into plasma.
"This suggests that cfDNA has the potential to map the tumor epigenome in real time and therefore can help uncover the regulatory landscape of cancer from plasma," Ramachandran said.
According to the study, these findings could lead to a genome-wide map for defining disease state, predicting treatment outcome and perhaps choosing the most effective cancer therapy. None of that is practical now due to the risks involved in obtaining tumor tissue.
"In our study, we leveraged an alternate means to obtain the same information…in a minimally invasive manner to define underlying disease biology," Kabos and Ramachandran wrote.
Both said the information gleaned from the DNA of cancer could be used to develop new therapies in the future. And the same plasma analysis used in breast cancer, they said, would likely work with other malignancies.
"Given that most tumors release cfDNA, we believe that our characterization of ER+ breast tumors using cfDNA transcription factor footprints represents the tip of the iceberg for characterizing tumor phenotypes from plasma and is applicable across disease states," said Ramachandran, CU Cancer Center member.
More information: Satyanarayan Rao et al, Transcription factor–nucleosome dynamics from plasma cfDNA identifies ER-driven states in breast cancer, Science Advances (2022). DOI: 10.1126/sciadv.abm4358
Journal information: Science Advances
Provided by CU Anschutz Medical Campus