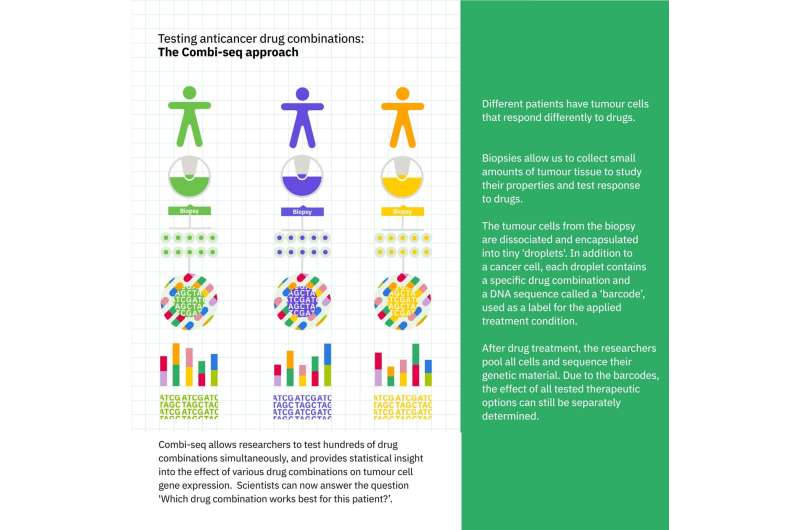
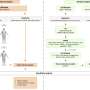
Combi-seq allows researchers to test hundreds of drug combinations simultaneously for their effect on the transcriptomes of cancer cells. Credit: Aleksandra Krolik/EMBL
Each year, around 10 million lives around the world are cut short by cancer. While the last century of research has seen the number of approved anticancer drugs grow by leaps and bounds, the sheer diversity of disease forms and patient responses often defeats treatment strategies. The next revolution in anti-cancer therapeutics is likely to arrive in the form of drug combinations tailored specifically to a patient's own, unique tumor cells.
A new study from the group of Christoph Merten, former group leader at EMBL Heidelberg and presently at the Swiss Federal Institute of Technology Lausanne (EPFL), proposes a new approach that will greatly increase the power of drug efficacy testing and pave the way for more personalized cancer therapy.
The study was carried out in collaboration with the group of Julio Saez-Rodriguez, Professor at the Medical Faculty of Heidelberg University, a Group Leader at EMBL—University Hospital Heidelberg Molecular Medicine Partnership Unit (MMPU), and former group leader at EMBL-EBI, who supervised the computational analysis, led by Bence Szalai.
Since every patient's tumor cells are different and continuously accumulate mutations, it is difficult to predict how they will react to a specific drug. The chance of tumor cells developing resistance to a particular drug is also high. For these reasons, combinations of two or more drugs are often considered more promising than single-drug chemotherapy. However, testing such combinations presents its own challenges.
Due to the limited amount of patient tissue, only a few drug combinations can be tested in the traditional approach. The results of testing are binary in nature, i.e. whether the tumor cells live or die upon treatment, so we can only answer the question "Is the patient resistant or responsive to this drug combination?" Credit: Aleksandra Krolik/EMBL
"When testing the effects of drugs, we are limited by the amount of tissue we can obtain from patient biopsies, which is generally low. This means that with conventional technology, it's impossible to screen hundreds of drug combinations for their effect on patient tumor cells," Merten said.
Due to this reliance on large tissue volumes, present technologies can only test a handful of drug conditions at a time. They also usually provide purely yes-or-no answers, e.g. whether the cells live or die after treatment.
However, Combi-seq—the new technique—overcomes this by cleverly using microfluidics—the precise control and manipulation of fluids with very miniaturized devices. Because of the low volume of liquids required, this allows researchers to carry out large-scale experiments with very small sample volumes. In 2018, Merten's and Saez-Rodriguez's groups employed microfluidics to test 56 anticancer drug combinations in cancer cells from patients.
The present technique, established by former EMBL Ph.D. student Lukas Mathur, takes this process even further, allowing researchers to test over 400 drug combinations with only a tiny amount of biological material.
On the contrary, Combi-seq allows researchers to test hundreds of drug combinations simultaneously, and provides statistical insight into the effect of various drug combinations on tumor cell gene expression. Scientists can now answer the question "Which drug combination works best for this patient?" Credit: Aleksandra Krolik/EMBL
Combi-seq works by precise microfluidic manipulation of cells in solution. The researchers used this to isolate cells in droplets, each of which was only around a tenth of a millimeter in diameter. In addition to a cancer cell, each droplet contained a specific drug combination and a DNA sequence called a "barcode," used as a label for the applied treatment condition. After 12 hours of treatment, the researchers could pool the cells, collect their genetic material for sequencing (identified by the "barcodes"), and analyze the results.
Not only does Combi-seq massively scale up how many treatment conditions can be tested simultaneously, it also allows scientists to gather accurate transcriptomics data from drug-exposed cells. It does so by incorporating next-generation sequencing into the workflow. Instead of just telling us whether a cell lives or dies after drug treatment, this method gives us a wealth of information about the cell's response that medical professionals might be able to draw upon while determining treatment strategies.
"Using such transcriptomics data, we can make statements about how the signaling pathways in the cell react to the drug or about which genes are up- or downregulated in response. This is so much more powerful than anything we have had previously," Merten said.
"Generating these datasets for different patients for a tumor type and applying advanced computational methods on them can improve our understanding of why drugs often do not work and ultimately improve patient care," said Julio Saez-Rodriguez.
The method is both powerful and versatile. "Our primary interest is in anticancer therapeutics," Merten said. "However, this technique can potentially be extended to any conditions where one needs to analyze the response of primary human cells to drugs in detail."
More information: L. Mathur et al, Combi-seq for multiplexed transcriptome-based profiling of drug combinations using deterministic barcoding in single-cell droplets, Nature Communications (2022). DOI: 10.1038/s41467-022-32197-0
Federica Eduati et al, A microfluidics platform for combinatorial drug screening on cancer biopsies, Nature Communications (2018). DOI: 10.1038/s41467-018-04919-w
Journal information: Nature Communications
Provided by European Molecular Biology Laboratory