Exposing the evolutionary weak spots of the human genome
Mutations can be good and bad. Sometimes they help an organism adapt and survive. Other times they are so harmful that an organism can't survive or reproduce. Cold Spring Harbor Laboratory (CSHL) Professor Adam Siepel's team has created a computer program that tracks the history of harmful mutations in the human genome throughout evolution.
They discovered parts of the genome are especially vulnerable to mutations, meaning any mutations in those regions can result in severe or lethal consequences. Their findings may help guide clinicians seeking the origins of serious genetic diseases.
Siepel's program is called ExtRaINSIGHT. It finds harmful mutations by looking for their absence. By random chance, every part of the human genome should have mutations but some have none. Siepel calls these places "ultraselected." When they occur, the mutations there can be deadly or drastically hurt the odds of reproducing. Siepel explains:
"If we look across a panel of a hundred thousand humans and we never see a mutation at a particular gene, that suggests that any mutation that did occur was so harmful, that anyone carrying that mutation died out from the population."
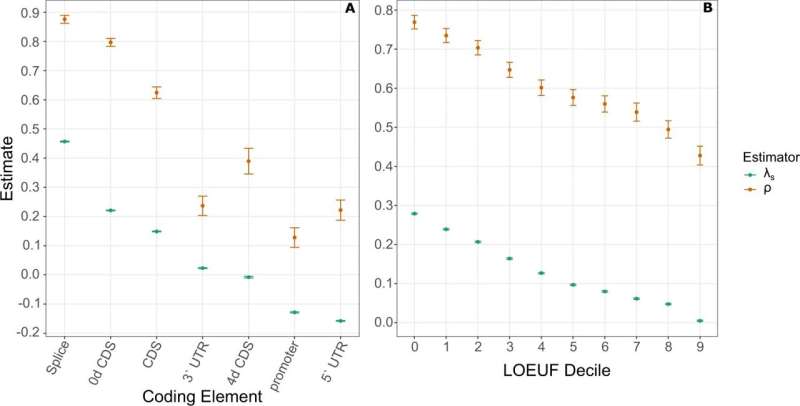
The team analyzed over 70,000 human genomes with ExtRaINSIGHT. They discovered that three parts of the genome have been extremely sensitive to mutations over generations. Of these, splice sites are the most sensitive. Splice sites help produce correct instructions for making proteins. Mutations here can have a huge impact on the odds of passing down genes, also known as fitness. They're linked to several diseases including spinal muscular atrophy, the leading genetic cause of death in infants and toddlers. Siepel says:
"If you see a mutation in a splice site, you better take it seriously. That mutation alone would reduce your fitness by 1 or 2%. That doesn't sound like very much, but that's a huge fitness effect. And if you had multiple of these, pretty soon your chance of passing on your genes might be close to zero."
Molecules called miRNA and central nervous system genes are also sensitive. "If you find a mutation in miRNA there's a good chance it's responsible for a genetic disease," Siepel says. "And because the nervous system is so complex and interconnected, it seems particularly sensitive to mutation."
The origins of many genetic diseases and conditions remain a mystery. Siepel hopes technology like ExtRaINSIGHT will help reveal their origins and guide diagnoses and future treatments. He also hopes his work will help further illustrate how mutations continue to shape the evolution of the human genome.
The research was published in Nature Communications.
More information: Noah Dukler et al, Extreme purifying selection against point mutations in the human genome, Nature Communications (2022). DOI: 10.1038/s41467-022-31872-6